Diagnosiscwp-140
cwp-140 时间:2021-04-30 阅读:()
AdvExpMedBiol–AdvancesinMicrobiology,InfectiousDiseasesandPublicHealth(2018)8:255–261https://doi.
org/10.
1007/978-3-319-72799-8#SpringerInternationalPublishingAG2018IndexAABCDmodel,82Activeimmunization,208,211–214mucosalimmunization,213–214parenteralimmunization,211,213vaccinestargetingsurfacecomponents,214–216vaccinestargetingtoxinsinanimalmodels,208–211inhumansandclinicaltrials,211–214Agardilution(AD)methodadvantages,142disadvantages,142,145Agarincorporation(AI),145,146Airbornesporetransmission,238Animalmodelassays,antibody-basedproductsmucosaladministration,204–205parenteraladministration,202–204Antibioticresistance,239geneticanalysis,138multi-drugresistance,142–144resistancemechanismscephalosporins,146–148chloramphenicol,147,152daxomicin,151uoroquinolones,147,149metronidazole,147,149MLSB,147–149rifamycins,147,151tetracycline,147,151–152vancomycin,147,149,151ribotype,138susceptibilityagardilution,142,145agarincorporation,145,146brothmicrodilution,145,146cephalosporins,138–140ciprooxacin,139clindamycin,138diskdiffusiontesting,146epsilometertest,142,145daxomicin,138uoroquinolones,138–140gatioxacin,139metronidazole,138–140moxioxacin,139phenotypictests,146rifamycins,138,140–142rifaximin,138vancomycin,138–140AntibioticseconomicburdenofClostridiumdifcileinfection,2effects,107–109FMT,177–178Antibody-basedproducts(AP),201mucosaladministration,204–205parenteraladministration,202–204passiveimmunizationstrategies,201–208Antibodyengineering,202Arginine-177,88Autoinducers(AI),98BBacteriophages,69,70,109–110Bezlotoxumab,124,206,207Binarytoxin,51,200Biolmadhesiveandinvasivefeatures,98adhesiveproperties,98alternativetherapeuticagents,109anti-biolmcompounds,109antibioticseffects,107–109bacteriophages,109–110CdiBs21,99cellswithspatiallocalisation,100CLSManalysis,99–102complexmultifactorialprocess,99eDNA,100EPSmatrix,99extracellularpolymericsubstances,97fermentationproles,98FESEManalysis,100–102formation,97–98geneticfactorsautoinducermolecules,105c-di-GMP,103–104CodY,104DegS/DegUhomologues,104iCmutant,104255Biolm(cont.
)F2locus,104Hfq,104–105immunogoldlabelling,103LexA,106pilA1–3pilA1transcripts,103S-layer,105smallnon-codingRNAs,104Spo0A-P,105T4P,102–103hypervirulentstrainR20291,99indirectimmunouorescenceanalysis,100Manukahoney,109MazE-MazFTAsystem,100metabolicandimmunologichomeostasis,98modeofgrowth,99outermostexosporiumlayer,100photodynamictherapy,110quorumsensing,98spo0Amutant,99TAsystems,100toxinsandspores,100virulencefactors,98invitroandinvivomodels,106–107Biogasplants,238Bis-(3'-5')-cyclicdimericguanosinemonophosphate(c-di-GMP),103BovineimmunoglobulinGconcentrate(BIC),204Brothmicrodilution,145,146CCaco-2,seeEnterocyte-likemodelCadazolid,124–125Calprotectin,35–37CbpA,199CDT,seeClostridiumdifciletransferaseCellcytotoxicityneutralizationassay(CCNA),28–29Cell-wallproteins,198–199Cephalosporins(CFs)mechanismsofresistance,146–148susceptibility,138–140Chaperones,88Chasertherapy,138Chemokines,40Chloramphenicol(CHL),147,152Ciprooxacin(CIP),139Clindamycin(CLI),138ClinicalandLaboratoryStandardsInstitute(CLSI),142Clostridiumdifcilecolonization,229–231Clostridiumdifcileguidelines,seeEuropeanSocietyofClinicalMicrobiologyandInfectiousDiseases(ESCMID)StudyGroupClostridiumdifcileinfection(CDI)activeimmunisation,126–127antibioticresistances(seeAntibioticresistances)bezlotoxumab,124biolm(seeBiolm)cadazolid,125clinicalmanifestation,98clinicalsigns,197incommunity,53–54comparativegenomics(seeComparativegenomics)diagnosis,179(seeDiagnosis)diagnosticmethods,249economicburden(seeEconomicburdenofClostridiumdifcileinfection)epidemiology,249–250,252ESCMIDguidelinesclinicalscenarios,119,120non-severeCDI,119recurrentCDI,121severeCDI,119–123Europeandiagnosticguidance,251Europeansurveyanti-CDItherapy,122–124Demographixsoftware,121Exceldatabase,121nationalguidelines,122,123farmanimals(seeFarmanimals)daxomicin,124healthcare,197healthcare-associatedinfections,178intestinalmicrobiota,198laboratoryinvestigation,249management,250–251metronidazole,124microbiomebasedtherapeuticsFMT,127–128NTCD,130–131RBX2660,128–129SER-109,129–130molecularsubtyping,250morbidityandmortality,197–198probiotics(seeProbiotics)prophylaxisDAV132,126ribaxamase,125–126ridinilazole,125SIGHTMnemonicprotocol,118surotomycin,124surveillance(seeSurveillance)treatment,198Clostridiumdifcileresearch,seeEuropeanSocietyofClinicalMicrobiologyandInfectiousDiseases(ESCMID)StudyGroupClostridiumdifciletransferase(CDT),200bindingcomponent,86bipartitecomposition,86cellularuptake,endocyticpathwaysfor,87–88chaperonesrole,88enzymecomponent,86–87lipolysis-stimulatedlipoproteinreceptor,87mode-of-action,88–89roleof,89Clostridiumdifciletyping,46–47Clostridiumdifcilevirulencefactors,198–201CoDIFy,125256IndexCodY,104Colonizationfactors,198,204,208Combined,repetitiveoligopeptides(CROP),78,80Community-acquiredCDI,6,8Companionanimals,ClostridiumdifcileComparativegenomehybridization(CGH),60Comparativegenomicscompletegenomesequence,60ne-scalecomparativegenomicapproaches,implementationof,60,61globalcomparativegenomicsCD630,virulenceandphenotypeof,66non-toxigenicstrains,66–67PaLocacquisitionandexchange,67PCRribotype027,63–64populationstructure,60–63reinfectionsvs.
relapses,65–66ribotype017,64transmissions,65moleculartypingmethods,59targetedcomparativegenomicsCRISPR/Cassystemsandphage-hostinteraction,69CRISPRdistributionanddiversity,70CRISPRmechanismandphysiology,69–70PaLocorganizationandevolution,67–69pathogenicitylocus,67ConfocalLaserScanningMicroscopy(CLSM),99–102CRISPRCRISPR/Cassystemsandphage-hostinteraction,69distributionanddiversity,70mechanismandphysiology,69–70Crohn'sdisease(CD),186Crypticclades,60CTimaging,40Cwp84protease,199Cwp66protein,199CXCL-5,40Cysteineproteasedomain,80–81DDegS-DegUtwo-componentsignaltransductionsystem,104Diagnosis,28alternativetestingstrategiescalprotectin,35–37CTimaging,40endoscopy,40–41faecalleukocytetest,39histopathology,41interleukinsandchemokines,40lactoferrin,37–39assaysavailability,28rapidassays,29recommendedtestingalgorithms,29–32referencetests,28–29repeattesting,33–34stoolsampleselection,32–33testingstrategy,consequencesof,34Diskdiffusiontesting,146Dysbiosis,161,162,167,234EEconomicburdenofClostridiumdifcileinfectionantibiotics,2community-acquiredCDI,6,8costs,9–10diagnosisandtreatment,advancesin,1–2healthcareand,2hospital-acquiredCDIcostdistribution,5–7costsbystudyandcountry,3lengthofstay,5,6primaryepisodes,2recurrentepisodes,4–5SouthernEurope,4WesternEurope,2–4meta-analysis,2pediatricpopulation,8–9silicoeconomicmodel,2Endoscopy,40–41Enterocyte-likemodel,166Enzymeimmunoassays(EIA),28,29Epsilometertest,142,145ESCMIDStudyGroupforClostridiumdifcile(ESGCD),246–247activitiesandachievements,249–251committeeof,247,248historyandorigins,247–248ESCMIDStudyGroups'competenciesinantimicrobialprescribingandstewardship(ESCAPS),251EuropeanCentreforDiseasePreventionandControl(ECDC),252EuropeanCommitteeonAntimicrobialSusceptibilityTesting(EUCAST),145EuropeanReferenceNetwork(ERN),252EuropeanSocietyofClinicalMicrobiologyandInfectiousDiseases(ESCMID)clinicalscenarios,119,120non-severeCDI,119recurrentCDI,121severeCDI,119–123EuropeanSocietyofClinicalMicrobiologyandInfectiousDiseases(ESCMID)StudyGroup,245–246EuropeanStudyGroup(ESGCD),21,246–251EuropeansurveillanceofCDIbenets,19–21collaborativeefforts,21diagnosingandmonitoringcases,16–18ECDClong-termsurveillancestrategy,targetsin,17epidemiology,14EuropeanStudyGroup,21feedbackof,16formativedocuments,21guidance,development,14–16laboratorybasedsurveillance,benetsof,23monitoring,capabilityandcapacityfor,18multi-countrysurveillanceprogram,21Index257EuropeansurveillanceofCDI(cont.
)over-archinglong-termsurveillancestrategy,22pre-requisitefor,19signicantreductions,23standardisation,21,23ExtracellularDNA(eDNA),99,100Extracellularmatrix(ECM)proteins,89Extracellularpolymericsubstances46(EPS),97FF-actin,88,89,171Faecalcalprotectin(FCP),35–37Faecallactoferrin(FL),37–39Faecalleukocytetest,39Farmanimals,228,229,239–240antimicrobialsusceptibility,232cattle,228andcolonization,231–232drugresistance,232factors,229–231goats,228gutmicrobiotacomposition,231infectionvs.
carriage,231pigs,228poultry,228prevalence,230sheep,228zoonotictransmission,228–229FbpA,199Fecalmicrobiotatransplantation(FMT),118,121,127–128antibiotics,177–178characteristics,180–182Clostridiumdifcileburden,178–179colonoscopy,183communityandhospital-acquired,184donorscreening,186efcacyandsafety,186enema,184futilityanalysis,183forIBD,178,186adverseevents,191characteristics,188–190clinicalpractice,191mortality,191medicaltreatment,177bynasoduodenaltube,184randomizedcontrolledtrials,179rCDItreatment,179,185treatmentstrategy,177–178upperGIdelivery,184Fidaxomicin(FDX)CDItreatment,124mechanismsofresistance,151susceptibility,138Flagellarproteins,199iCgene,104Fluoroquinoloneresistant,64Fluoroquinolones(FQs)mechanismsofresistance,147,149susceptibility,138–140FMT,seeFecalmicrobiotatransplantationFood,235contaminatedmeats,235–236humaninfection,239–240production,229seafood,236vegetables,236Fragmentsantibody,204GGatioxacin(GAT),139GermanClinicalMicrobiomeStudyGroup(GCMSG),183Glucosylation,84Glucosyltransferasedomain,80Glutamatedehydrogenaseenzymeimmunoassay(GDHEIA),29,30Gutmicrobiota,162,177,231pathogenesis,178severity,178HHeatshockprotein,GroEL,199Hfqprotein,104–105Histopathology,41Horses,ClostridiumdifcileHospital-acquiredCDIcostdistribution,5–7costsbystudyandcountry,3lengthofstay,5,6primaryepisodes,2recurrentepisodes,4–5SouthernEurope,4WesternEurope,2–4Hosthumoralimmuneresponse,200–201HT29models,166Humanassaysandclinicaltrials,205–208mucosaladministration,AP,207–208parenteraladministration,AP,205–207passiveimmunotherapy,205Human"colonic"model,165IIBD,seeInammatoryboweldiseaseICDS,seeInternationalClostridiumdifcileSymposiumIL8,40Immunizationactiveimmunization,211–214immunization:mucosalimmunization,213–214immunization:parenteralimmunization,211,213vaccinestargetingsurfacecomponents,214–216vaccinestargetingtoxinsvaccinestargetingtoxins:inanimalmodels,208–211258Indexvaccinestargetingtoxins:inhumansandclinicaltrials,211–214passiveanimalmodelsassays,202–205antibodyengineering,202assaysinhumansandclinicaltrials,205–208bioavailability,201inclinicaldevelopment,206lowimmunogenicity,201surfaceproteinsandcolonizationfactors,198–199toxins,199–200Immunoglobulintherapy,122Indirectimmunouorescenceanalysis,100Inammatoryboweldisease(IBD),178,186–187adverseevents,191characteristics,188–190clinicalpractice,191mortality,191Inositolhexakisphosphate(InsP6),80Interleukins,40InternationalClostridiumdifcileSymposium(ICDS),248,251Intestinalpathogen,197Invitroexperimentalmodels,165,166Iotatoxin(Ib),86IStrons,69LLactoferrin,37–39Largeclostridialtoxins(LCTs),78LexA,106Lipolysis-stimulatedlipoproteinreceptor,87Livestockanimals,ClostridiumdifcileLocusofpathogenicity(PaLoc),46–47MMAbs,seeMonoclonalantibodiesMacrolide-lincosamide-streptograminB(MLSB),147–149Manukahoney,109MazE-MazFTAsystem,100MDR,seeMulti-drugresistanceMetronidazole(MTZ)CDItreatment,124Clostridiumdifcilemechanismsofresistance,147,149Clostridiumdifcilesusceptibility,138–140Microbialculturingmodels,165MLSB,seeMacrolide-lincosamide-streptograminBMobilegeneticelements(MGE),69Monoclonalantibodies(MAbs)inhumanassay,205–207againsttoxins,animalmodelassays,202–204Moxioxacin(MXF),139MTZ,seeMetronidazoleMucosaladministrationanimalmodelassays,204–205humanassays,207–208Mucosalimmunizationactiveimmunizationanimalmodels,210–211,215–216humansandclinicaltrials,213–214toTcdB,201Multi-drugresistance(MDR),142–144Multilocussequencetyping(MLST),60Multilocusvariable-numbertandemrepeatanalysis(MLVA),47NNAAT,seeNucleicacidamplicationtestNasoduodenaltube,184Next-generationsequencing(NGS),60Non-humanClostridiumdifcilereservoirsincompanionanimals,232–233inenvironment,236–239infarmanimals,228–232infood,235–236inhorses,233–234inwildanimals,234Nontoxicenvironmentalstrains,239Non-toxigenicClostridiumdifcile(NTCD),130–131NTCD,seeNon-toxigenicClostridiumdifcileNucleicacidamplicationtest(NAAT),28–30,32–34PParenteraladministration,passiveimmunizationanimalmodelassays,202–204humanassays,205–207Parenteralimmunization,activeimmunizationanimalmodels,208–210,214–215humansandclinicaltrials,211,213Passiveimmunizationanimalmodelsassays,202–205antibodyengineering,202bioavailability,201inclinicaldevelopment,206humansandclinicaltrials,assaysin,205–208lowimmunogenicity,201PCRribotyping,46,63A+B-CDT-unusualprole,emergingstrainswith,52antibioticresistance,138binarytoxin,50–53CA-CDI,epidemiologyof,53circulatingandemerging,characteristics,53Clostridiumdifcilediscriminatorypower,47geographicaldistribution,49globaldistribution,47–49PCRribotype017(RT017),51–52,64PCRribotype018(RT018),51PCRribotype027(RT027),63–64PCRribotype033(RT033),51PCRribotype078(RT078),50PCRribotype106(RT102),52PCRribotype126(RT126),50–51PCRribotype176(RT176),49–50PCRribotype244(RT244),52Pediatricpopulation,economicburdenofCDI,8–9Index259Pets,ClostridiumdifcilePhage-hostinteraction,69Phenotypictests,146Photodynamictherapy,110Polyclonalantibodiesagainstsurfaceproteins,204againsttoxins,202Polysaccharides(PS),99,100,199Polyvalentimmunoglobulins,205Post-antibioticdiarrhea,197Probioticsalternativetherapeuticoptions,162,163antibiotic-associateddiarrhea,161–162clinicalefcacy,163–165denition,162dysbiosisofmicrobiota,162enterocyte-likemodel,166gutmicrobiota,165hamstermodel,166HT29models,166human"colonic"model,165invitroexperimentalmodels,165,166mechanismsofactionanti-toxinactivity,168,169,172Clostridiumdifciletoxinactivity,169–170competitiveexclusion/co-aggregation,167–168immuneresponsemodulation,167inhibitingpathogenicmicroorganisms,167–469innateandadaptiveimmuneresponse,171intestinalmicrobiota,167productionofanti-microbialcompounds,168reinforcementoftheintestinalbarrier,167,168,170,171microbialculturingmodels,165ribotypes,162RTCAtechnology,166toxins,162ProphylaxisDAV132,126ribaxamase,125–126Pseudomembranouscolitis(PMC),40,197Pulsed-eldgelelectrophoresis(PFGE),32,47PUNCHOpenLabelstudy,129Pyrin,85Pyroptosis,85QQuorumsensing(QS),98,105,169RRacQ61Lmutant,84RBX2660,128–129RecurrentCDI(rCDI),178Reinfections,65–66Relapse,65–66Researchprojects,Clostridiumdifcile,seeEuropeanSocietyofClinicalMicrobiologyandInfectiousDiseases(ESCMID)StudyGroupRFs,seeRifamycinsRhoproteins,84Ridinilazole,125Rifamycins(RFs),140–142mechanismsofresistance,147,151susceptibility,138,140–142Rifaximin(RFX),138RTCAtechnology,166SSecreted-zincmetalloprotease,199Septins,89SER-109,129–130SevereCDI,denitionandtreatment,119–123Short-triptoxins,83SIGHTMnemonicprotocol,118S-layerproteins(SLPs),105,198Smallnon-codingRNAs(sRNAs),104Soil,ClostridiumdifcileSpacers,69Spo0A-P,105Standardisednationalsurveillanceprogrammes,21Stoolassays,16Straintyping,238Sumoftandemrepeatnumberdifferences(STRD),47SurfaceproteinsCbpA,199cell-wallproteins,198–199andcolonizationfactors,198–199FbpA,199agellarproteins,199polysaccharides,199Surotomycin,124Surveillancebenets,19–21collaborativeefforts,21diagnosingandmonitoringcases,16–18ECDClong-termsurveillancestrategy,targetsin,17epidemiology,14EuropeanStudyGroup,21feedbackof,16formativedocuments,21guidance,development,14–16laboratorybasedsurveillance,benetsof,23monitoring,capabilityandcapacityfor,18multi-countrysurveillanceprogram,21over-archinglong-termsurveillancestrategy,22pre-requisitefor,19signicantreductions,23standardisation,21,23TTetracycline(TET),147,151–152Tn6218,68–69Toll-likereceptor4(TLR4),198Toxigenicculture(TC),28–29,32ToxinAandB,28,78,179,199–200ABCDmodel,82bindinganduptakecellularuptake,endocyticpathwaysfor,82–83260Indexcytopathologicaleffects,84glucosyltransferasedomain,83hostreceptors,82CDTbindingcomponent,86bipartitecomposition,86cellularuptake,endocyticpathwaysfor,87–88chaperonesrole,88enzymecomponent,86–87lipolysis-stimulatedlipoproteinreceptor,87mode-of-action,88–89roleof,89CROPdomain,78,80cysteineproteasedomain,80–81glucosyltransferasedomain,80importanceof,85–86mode-of-action,83–85modularcomposition,78receptor-bindingdomain,81–82short-triptoxins,83translocationdomain,81uptakeprocessandmode-of-action,78,79vaccinestargetinginanimalmodels,208–211inclinicaldevelopment,212–213inhumansandclinicaltrials,211–214Toxin-antitoxin(TA)systems,100Toxinotype,47ToxinotypeXIstrains,51ToxinsA/Benzymeimmunoassay(ToxA/BEIA),28–34Transmission,animalsandenvironment,234–235Triple-stagehumangutmodel,108TypeIVpili(T4P),102–103VVaccinationinducedpotentantitoxin,213targetingsurfacecomponentsinanimalmodels,214–216targetingtoxinsinanimalmodels,208–211inclinicaldevelopment,212–213inhumansandclinicaltrials,211–214Vancomycin(VAN)mechanismsofresistance,147,149,151susceptibility,138–140WWater,ClostridiumdifcileWholegenomesequencing(WGS),47,65Wildanimals,ClostridiumdifcileIndex261
org/10.
1007/978-3-319-72799-8#SpringerInternationalPublishingAG2018IndexAABCDmodel,82Activeimmunization,208,211–214mucosalimmunization,213–214parenteralimmunization,211,213vaccinestargetingsurfacecomponents,214–216vaccinestargetingtoxinsinanimalmodels,208–211inhumansandclinicaltrials,211–214Agardilution(AD)methodadvantages,142disadvantages,142,145Agarincorporation(AI),145,146Airbornesporetransmission,238Animalmodelassays,antibody-basedproductsmucosaladministration,204–205parenteraladministration,202–204Antibioticresistance,239geneticanalysis,138multi-drugresistance,142–144resistancemechanismscephalosporins,146–148chloramphenicol,147,152daxomicin,151uoroquinolones,147,149metronidazole,147,149MLSB,147–149rifamycins,147,151tetracycline,147,151–152vancomycin,147,149,151ribotype,138susceptibilityagardilution,142,145agarincorporation,145,146brothmicrodilution,145,146cephalosporins,138–140ciprooxacin,139clindamycin,138diskdiffusiontesting,146epsilometertest,142,145daxomicin,138uoroquinolones,138–140gatioxacin,139metronidazole,138–140moxioxacin,139phenotypictests,146rifamycins,138,140–142rifaximin,138vancomycin,138–140AntibioticseconomicburdenofClostridiumdifcileinfection,2effects,107–109FMT,177–178Antibody-basedproducts(AP),201mucosaladministration,204–205parenteraladministration,202–204passiveimmunizationstrategies,201–208Antibodyengineering,202Arginine-177,88Autoinducers(AI),98BBacteriophages,69,70,109–110Bezlotoxumab,124,206,207Binarytoxin,51,200Biolmadhesiveandinvasivefeatures,98adhesiveproperties,98alternativetherapeuticagents,109anti-biolmcompounds,109antibioticseffects,107–109bacteriophages,109–110CdiBs21,99cellswithspatiallocalisation,100CLSManalysis,99–102complexmultifactorialprocess,99eDNA,100EPSmatrix,99extracellularpolymericsubstances,97fermentationproles,98FESEManalysis,100–102formation,97–98geneticfactorsautoinducermolecules,105c-di-GMP,103–104CodY,104DegS/DegUhomologues,104iCmutant,104255Biolm(cont.
)F2locus,104Hfq,104–105immunogoldlabelling,103LexA,106pilA1–3pilA1transcripts,103S-layer,105smallnon-codingRNAs,104Spo0A-P,105T4P,102–103hypervirulentstrainR20291,99indirectimmunouorescenceanalysis,100Manukahoney,109MazE-MazFTAsystem,100metabolicandimmunologichomeostasis,98modeofgrowth,99outermostexosporiumlayer,100photodynamictherapy,110quorumsensing,98spo0Amutant,99TAsystems,100toxinsandspores,100virulencefactors,98invitroandinvivomodels,106–107Biogasplants,238Bis-(3'-5')-cyclicdimericguanosinemonophosphate(c-di-GMP),103BovineimmunoglobulinGconcentrate(BIC),204Brothmicrodilution,145,146CCaco-2,seeEnterocyte-likemodelCadazolid,124–125Calprotectin,35–37CbpA,199CDT,seeClostridiumdifciletransferaseCellcytotoxicityneutralizationassay(CCNA),28–29Cell-wallproteins,198–199Cephalosporins(CFs)mechanismsofresistance,146–148susceptibility,138–140Chaperones,88Chasertherapy,138Chemokines,40Chloramphenicol(CHL),147,152Ciprooxacin(CIP),139Clindamycin(CLI),138ClinicalandLaboratoryStandardsInstitute(CLSI),142Clostridiumdifcilecolonization,229–231Clostridiumdifcileguidelines,seeEuropeanSocietyofClinicalMicrobiologyandInfectiousDiseases(ESCMID)StudyGroupClostridiumdifcileinfection(CDI)activeimmunisation,126–127antibioticresistances(seeAntibioticresistances)bezlotoxumab,124biolm(seeBiolm)cadazolid,125clinicalmanifestation,98clinicalsigns,197incommunity,53–54comparativegenomics(seeComparativegenomics)diagnosis,179(seeDiagnosis)diagnosticmethods,249economicburden(seeEconomicburdenofClostridiumdifcileinfection)epidemiology,249–250,252ESCMIDguidelinesclinicalscenarios,119,120non-severeCDI,119recurrentCDI,121severeCDI,119–123Europeandiagnosticguidance,251Europeansurveyanti-CDItherapy,122–124Demographixsoftware,121Exceldatabase,121nationalguidelines,122,123farmanimals(seeFarmanimals)daxomicin,124healthcare,197healthcare-associatedinfections,178intestinalmicrobiota,198laboratoryinvestigation,249management,250–251metronidazole,124microbiomebasedtherapeuticsFMT,127–128NTCD,130–131RBX2660,128–129SER-109,129–130molecularsubtyping,250morbidityandmortality,197–198probiotics(seeProbiotics)prophylaxisDAV132,126ribaxamase,125–126ridinilazole,125SIGHTMnemonicprotocol,118surotomycin,124surveillance(seeSurveillance)treatment,198Clostridiumdifcileresearch,seeEuropeanSocietyofClinicalMicrobiologyandInfectiousDiseases(ESCMID)StudyGroupClostridiumdifciletransferase(CDT),200bindingcomponent,86bipartitecomposition,86cellularuptake,endocyticpathwaysfor,87–88chaperonesrole,88enzymecomponent,86–87lipolysis-stimulatedlipoproteinreceptor,87mode-of-action,88–89roleof,89Clostridiumdifciletyping,46–47Clostridiumdifcilevirulencefactors,198–201CoDIFy,125256IndexCodY,104Colonizationfactors,198,204,208Combined,repetitiveoligopeptides(CROP),78,80Community-acquiredCDI,6,8Companionanimals,ClostridiumdifcileComparativegenomehybridization(CGH),60Comparativegenomicscompletegenomesequence,60ne-scalecomparativegenomicapproaches,implementationof,60,61globalcomparativegenomicsCD630,virulenceandphenotypeof,66non-toxigenicstrains,66–67PaLocacquisitionandexchange,67PCRribotype027,63–64populationstructure,60–63reinfectionsvs.
relapses,65–66ribotype017,64transmissions,65moleculartypingmethods,59targetedcomparativegenomicsCRISPR/Cassystemsandphage-hostinteraction,69CRISPRdistributionanddiversity,70CRISPRmechanismandphysiology,69–70PaLocorganizationandevolution,67–69pathogenicitylocus,67ConfocalLaserScanningMicroscopy(CLSM),99–102CRISPRCRISPR/Cassystemsandphage-hostinteraction,69distributionanddiversity,70mechanismandphysiology,69–70Crohn'sdisease(CD),186Crypticclades,60CTimaging,40Cwp84protease,199Cwp66protein,199CXCL-5,40Cysteineproteasedomain,80–81DDegS-DegUtwo-componentsignaltransductionsystem,104Diagnosis,28alternativetestingstrategiescalprotectin,35–37CTimaging,40endoscopy,40–41faecalleukocytetest,39histopathology,41interleukinsandchemokines,40lactoferrin,37–39assaysavailability,28rapidassays,29recommendedtestingalgorithms,29–32referencetests,28–29repeattesting,33–34stoolsampleselection,32–33testingstrategy,consequencesof,34Diskdiffusiontesting,146Dysbiosis,161,162,167,234EEconomicburdenofClostridiumdifcileinfectionantibiotics,2community-acquiredCDI,6,8costs,9–10diagnosisandtreatment,advancesin,1–2healthcareand,2hospital-acquiredCDIcostdistribution,5–7costsbystudyandcountry,3lengthofstay,5,6primaryepisodes,2recurrentepisodes,4–5SouthernEurope,4WesternEurope,2–4meta-analysis,2pediatricpopulation,8–9silicoeconomicmodel,2Endoscopy,40–41Enterocyte-likemodel,166Enzymeimmunoassays(EIA),28,29Epsilometertest,142,145ESCMIDStudyGroupforClostridiumdifcile(ESGCD),246–247activitiesandachievements,249–251committeeof,247,248historyandorigins,247–248ESCMIDStudyGroups'competenciesinantimicrobialprescribingandstewardship(ESCAPS),251EuropeanCentreforDiseasePreventionandControl(ECDC),252EuropeanCommitteeonAntimicrobialSusceptibilityTesting(EUCAST),145EuropeanReferenceNetwork(ERN),252EuropeanSocietyofClinicalMicrobiologyandInfectiousDiseases(ESCMID)clinicalscenarios,119,120non-severeCDI,119recurrentCDI,121severeCDI,119–123EuropeanSocietyofClinicalMicrobiologyandInfectiousDiseases(ESCMID)StudyGroup,245–246EuropeanStudyGroup(ESGCD),21,246–251EuropeansurveillanceofCDIbenets,19–21collaborativeefforts,21diagnosingandmonitoringcases,16–18ECDClong-termsurveillancestrategy,targetsin,17epidemiology,14EuropeanStudyGroup,21feedbackof,16formativedocuments,21guidance,development,14–16laboratorybasedsurveillance,benetsof,23monitoring,capabilityandcapacityfor,18multi-countrysurveillanceprogram,21Index257EuropeansurveillanceofCDI(cont.
)over-archinglong-termsurveillancestrategy,22pre-requisitefor,19signicantreductions,23standardisation,21,23ExtracellularDNA(eDNA),99,100Extracellularmatrix(ECM)proteins,89Extracellularpolymericsubstances46(EPS),97FF-actin,88,89,171Faecalcalprotectin(FCP),35–37Faecallactoferrin(FL),37–39Faecalleukocytetest,39Farmanimals,228,229,239–240antimicrobialsusceptibility,232cattle,228andcolonization,231–232drugresistance,232factors,229–231goats,228gutmicrobiotacomposition,231infectionvs.
carriage,231pigs,228poultry,228prevalence,230sheep,228zoonotictransmission,228–229FbpA,199Fecalmicrobiotatransplantation(FMT),118,121,127–128antibiotics,177–178characteristics,180–182Clostridiumdifcileburden,178–179colonoscopy,183communityandhospital-acquired,184donorscreening,186efcacyandsafety,186enema,184futilityanalysis,183forIBD,178,186adverseevents,191characteristics,188–190clinicalpractice,191mortality,191medicaltreatment,177bynasoduodenaltube,184randomizedcontrolledtrials,179rCDItreatment,179,185treatmentstrategy,177–178upperGIdelivery,184Fidaxomicin(FDX)CDItreatment,124mechanismsofresistance,151susceptibility,138Flagellarproteins,199iCgene,104Fluoroquinoloneresistant,64Fluoroquinolones(FQs)mechanismsofresistance,147,149susceptibility,138–140FMT,seeFecalmicrobiotatransplantationFood,235contaminatedmeats,235–236humaninfection,239–240production,229seafood,236vegetables,236Fragmentsantibody,204GGatioxacin(GAT),139GermanClinicalMicrobiomeStudyGroup(GCMSG),183Glucosylation,84Glucosyltransferasedomain,80Glutamatedehydrogenaseenzymeimmunoassay(GDHEIA),29,30Gutmicrobiota,162,177,231pathogenesis,178severity,178HHeatshockprotein,GroEL,199Hfqprotein,104–105Histopathology,41Horses,ClostridiumdifcileHospital-acquiredCDIcostdistribution,5–7costsbystudyandcountry,3lengthofstay,5,6primaryepisodes,2recurrentepisodes,4–5SouthernEurope,4WesternEurope,2–4Hosthumoralimmuneresponse,200–201HT29models,166Humanassaysandclinicaltrials,205–208mucosaladministration,AP,207–208parenteraladministration,AP,205–207passiveimmunotherapy,205Human"colonic"model,165IIBD,seeInammatoryboweldiseaseICDS,seeInternationalClostridiumdifcileSymposiumIL8,40Immunizationactiveimmunization,211–214immunization:mucosalimmunization,213–214immunization:parenteralimmunization,211,213vaccinestargetingsurfacecomponents,214–216vaccinestargetingtoxinsvaccinestargetingtoxins:inanimalmodels,208–211258Indexvaccinestargetingtoxins:inhumansandclinicaltrials,211–214passiveanimalmodelsassays,202–205antibodyengineering,202assaysinhumansandclinicaltrials,205–208bioavailability,201inclinicaldevelopment,206lowimmunogenicity,201surfaceproteinsandcolonizationfactors,198–199toxins,199–200Immunoglobulintherapy,122Indirectimmunouorescenceanalysis,100Inammatoryboweldisease(IBD),178,186–187adverseevents,191characteristics,188–190clinicalpractice,191mortality,191Inositolhexakisphosphate(InsP6),80Interleukins,40InternationalClostridiumdifcileSymposium(ICDS),248,251Intestinalpathogen,197Invitroexperimentalmodels,165,166Iotatoxin(Ib),86IStrons,69LLactoferrin,37–39Largeclostridialtoxins(LCTs),78LexA,106Lipolysis-stimulatedlipoproteinreceptor,87Livestockanimals,ClostridiumdifcileLocusofpathogenicity(PaLoc),46–47MMAbs,seeMonoclonalantibodiesMacrolide-lincosamide-streptograminB(MLSB),147–149Manukahoney,109MazE-MazFTAsystem,100MDR,seeMulti-drugresistanceMetronidazole(MTZ)CDItreatment,124Clostridiumdifcilemechanismsofresistance,147,149Clostridiumdifcilesusceptibility,138–140Microbialculturingmodels,165MLSB,seeMacrolide-lincosamide-streptograminBMobilegeneticelements(MGE),69Monoclonalantibodies(MAbs)inhumanassay,205–207againsttoxins,animalmodelassays,202–204Moxioxacin(MXF),139MTZ,seeMetronidazoleMucosaladministrationanimalmodelassays,204–205humanassays,207–208Mucosalimmunizationactiveimmunizationanimalmodels,210–211,215–216humansandclinicaltrials,213–214toTcdB,201Multi-drugresistance(MDR),142–144Multilocussequencetyping(MLST),60Multilocusvariable-numbertandemrepeatanalysis(MLVA),47NNAAT,seeNucleicacidamplicationtestNasoduodenaltube,184Next-generationsequencing(NGS),60Non-humanClostridiumdifcilereservoirsincompanionanimals,232–233inenvironment,236–239infarmanimals,228–232infood,235–236inhorses,233–234inwildanimals,234Nontoxicenvironmentalstrains,239Non-toxigenicClostridiumdifcile(NTCD),130–131NTCD,seeNon-toxigenicClostridiumdifcileNucleicacidamplicationtest(NAAT),28–30,32–34PParenteraladministration,passiveimmunizationanimalmodelassays,202–204humanassays,205–207Parenteralimmunization,activeimmunizationanimalmodels,208–210,214–215humansandclinicaltrials,211,213Passiveimmunizationanimalmodelsassays,202–205antibodyengineering,202bioavailability,201inclinicaldevelopment,206humansandclinicaltrials,assaysin,205–208lowimmunogenicity,201PCRribotyping,46,63A+B-CDT-unusualprole,emergingstrainswith,52antibioticresistance,138binarytoxin,50–53CA-CDI,epidemiologyof,53circulatingandemerging,characteristics,53Clostridiumdifcilediscriminatorypower,47geographicaldistribution,49globaldistribution,47–49PCRribotype017(RT017),51–52,64PCRribotype018(RT018),51PCRribotype027(RT027),63–64PCRribotype033(RT033),51PCRribotype078(RT078),50PCRribotype106(RT102),52PCRribotype126(RT126),50–51PCRribotype176(RT176),49–50PCRribotype244(RT244),52Pediatricpopulation,economicburdenofCDI,8–9Index259Pets,ClostridiumdifcilePhage-hostinteraction,69Phenotypictests,146Photodynamictherapy,110Polyclonalantibodiesagainstsurfaceproteins,204againsttoxins,202Polysaccharides(PS),99,100,199Polyvalentimmunoglobulins,205Post-antibioticdiarrhea,197Probioticsalternativetherapeuticoptions,162,163antibiotic-associateddiarrhea,161–162clinicalefcacy,163–165denition,162dysbiosisofmicrobiota,162enterocyte-likemodel,166gutmicrobiota,165hamstermodel,166HT29models,166human"colonic"model,165invitroexperimentalmodels,165,166mechanismsofactionanti-toxinactivity,168,169,172Clostridiumdifciletoxinactivity,169–170competitiveexclusion/co-aggregation,167–168immuneresponsemodulation,167inhibitingpathogenicmicroorganisms,167–469innateandadaptiveimmuneresponse,171intestinalmicrobiota,167productionofanti-microbialcompounds,168reinforcementoftheintestinalbarrier,167,168,170,171microbialculturingmodels,165ribotypes,162RTCAtechnology,166toxins,162ProphylaxisDAV132,126ribaxamase,125–126Pseudomembranouscolitis(PMC),40,197Pulsed-eldgelelectrophoresis(PFGE),32,47PUNCHOpenLabelstudy,129Pyrin,85Pyroptosis,85QQuorumsensing(QS),98,105,169RRacQ61Lmutant,84RBX2660,128–129RecurrentCDI(rCDI),178Reinfections,65–66Relapse,65–66Researchprojects,Clostridiumdifcile,seeEuropeanSocietyofClinicalMicrobiologyandInfectiousDiseases(ESCMID)StudyGroupRFs,seeRifamycinsRhoproteins,84Ridinilazole,125Rifamycins(RFs),140–142mechanismsofresistance,147,151susceptibility,138,140–142Rifaximin(RFX),138RTCAtechnology,166SSecreted-zincmetalloprotease,199Septins,89SER-109,129–130SevereCDI,denitionandtreatment,119–123Short-triptoxins,83SIGHTMnemonicprotocol,118S-layerproteins(SLPs),105,198Smallnon-codingRNAs(sRNAs),104Soil,ClostridiumdifcileSpacers,69Spo0A-P,105Standardisednationalsurveillanceprogrammes,21Stoolassays,16Straintyping,238Sumoftandemrepeatnumberdifferences(STRD),47SurfaceproteinsCbpA,199cell-wallproteins,198–199andcolonizationfactors,198–199FbpA,199agellarproteins,199polysaccharides,199Surotomycin,124Surveillancebenets,19–21collaborativeefforts,21diagnosingandmonitoringcases,16–18ECDClong-termsurveillancestrategy,targetsin,17epidemiology,14EuropeanStudyGroup,21feedbackof,16formativedocuments,21guidance,development,14–16laboratorybasedsurveillance,benetsof,23monitoring,capabilityandcapacityfor,18multi-countrysurveillanceprogram,21over-archinglong-termsurveillancestrategy,22pre-requisitefor,19signicantreductions,23standardisation,21,23TTetracycline(TET),147,151–152Tn6218,68–69Toll-likereceptor4(TLR4),198Toxigenicculture(TC),28–29,32ToxinAandB,28,78,179,199–200ABCDmodel,82bindinganduptakecellularuptake,endocyticpathwaysfor,82–83260Indexcytopathologicaleffects,84glucosyltransferasedomain,83hostreceptors,82CDTbindingcomponent,86bipartitecomposition,86cellularuptake,endocyticpathwaysfor,87–88chaperonesrole,88enzymecomponent,86–87lipolysis-stimulatedlipoproteinreceptor,87mode-of-action,88–89roleof,89CROPdomain,78,80cysteineproteasedomain,80–81glucosyltransferasedomain,80importanceof,85–86mode-of-action,83–85modularcomposition,78receptor-bindingdomain,81–82short-triptoxins,83translocationdomain,81uptakeprocessandmode-of-action,78,79vaccinestargetinginanimalmodels,208–211inclinicaldevelopment,212–213inhumansandclinicaltrials,211–214Toxin-antitoxin(TA)systems,100Toxinotype,47ToxinotypeXIstrains,51ToxinsA/Benzymeimmunoassay(ToxA/BEIA),28–34Transmission,animalsandenvironment,234–235Triple-stagehumangutmodel,108TypeIVpili(T4P),102–103VVaccinationinducedpotentantitoxin,213targetingsurfacecomponentsinanimalmodels,214–216targetingtoxinsinanimalmodels,208–211inclinicaldevelopment,212–213inhumansandclinicaltrials,211–214Vancomycin(VAN)mechanismsofresistance,147,149,151susceptibility,138–140WWater,ClostridiumdifcileWholegenomesequencing(WGS),47,65Wildanimals,ClostridiumdifcileIndex261
- Diagnosiscwp-140相关文档
- Isolationcwp-140
- 编号:AC-137-CA-2015-XX
- VRRMcwp-140
- 船舶cwp-140
- 起子cwp-140
- 三星cwp-140
CloudCone2核KVM美国洛杉矶MC机房机房2.89美元/月,美国洛杉矶MC机房KVM虚拟架构2核1.5G内存1Gbps带宽,国外便宜美国VPS七月特价优惠
近日CloudCone发布了七月的特价便宜优惠VPS云服务器产品,KVM虚拟架构,性价比最高的为2核心1.5G内存1Gbps带宽5TB月流量,2.89美元/月,稳定性还是非常不错的,有需要国外便宜VPS云服务器的朋友可以关注一下。CloudCone怎么样?CloudCone服务器好不好?CloudCone值不值得购买?CloudCone是一家成立于2017年的美国服务器提供商,国外实力大厂,自己开...
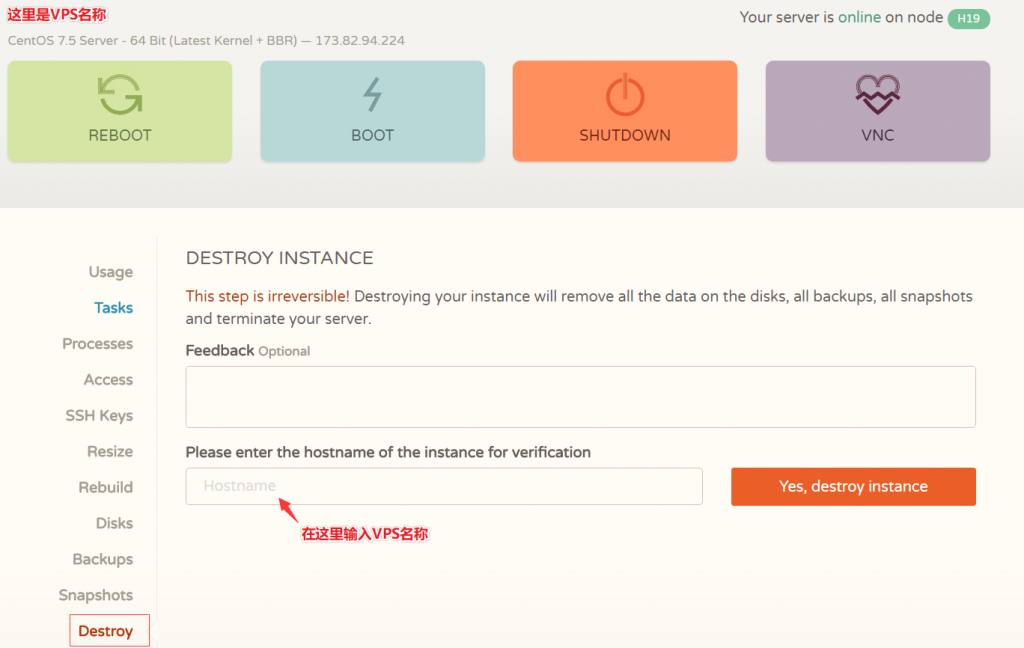
企鹅小屋:垃圾服务商有跑路风险,站长注意转移备份数据!
企鹅小屋:垃圾服务商有跑路风险!企鹅不允许你二次工单的,二次提交工单直接关服务器,再严重就封号,意思是你提交工单要小心,别因为提交工单被干了账号!前段时间,就有站长说企鹅小屋要跑路了,站长不太相信,本站平台已经为企鹅小屋推荐了几千元的业绩,CPS返利达182.67CNY。然后,站长通过企鹅小屋后台申请提现,提现申请至今已经有20几天,企鹅小屋也没有转账。然后,搞笑的一幕出现了:平台账号登录不上提示...
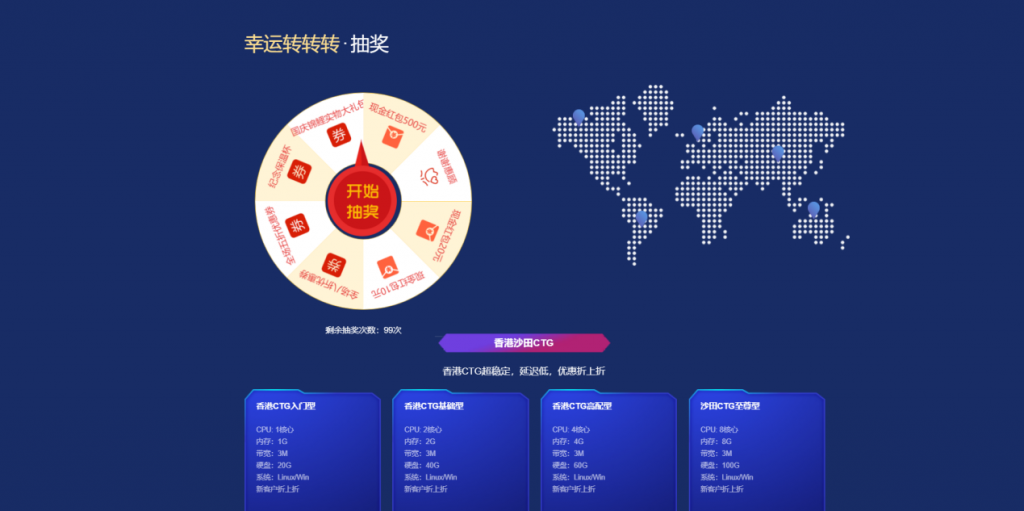
美国G口/香港CTG/美国T级超防云/物理机/CDN大促销 1核 1G 24元/月
[六一云迎国庆]转盘活动实物礼品美国G口/香港CTG/美国T级超防云/物理机/CDN大促销六一云 成立于2018年,归属于西安六一网络科技有限公司,是一家国内正规持有IDC ISP CDN IRCS电信经营许可证书的老牌商家。大陆持证公司受大陆各部门监管不好用支持退款退现,再也不怕被割韭菜了!主要业务有:国内高防云,美国高防云,美国cera大带宽,香港CTG,香港沙田CN2,海外站群服务,物理机,...
cwp-140为你推荐
-
strategicsns新闻联播网易yeahthinkphpthinkphp与PHP的差别,怎么查看thinkphp编写的系统?wordpress模板我在wordpress模板下载了一套模板,做了www.xuanqianbao.com这个站,模板的原站是www.rrzdm.com.一样的模板,我在文件在插入图片,却不能在首页显示,他的网站却可以.直outlookexpress家里电脑老是弹出“outlook express”这个东西,怎么除去啊?cuteftp什么是 CuteFtp Flashfxp Leapftp FlashGet三友网三友有机硅是不是国企,待遇如何?现在花钱去是不是值得?小型汽车网上自主编号申请成都新车上牌办理流程和办理条件是如何的青岛网通测速网通,联通,长城这三个宽带哪个网速最快?我是青岛的灌水机谁知道哪个好点的灌水机的地址?