branchesya
wx qq com yao 时间:2021-03-03 阅读:()
HUMANMUTATIONMutationinBrief#845(2005)OnlineMUTATIONINBRIEF2005WILEY-LISS,INC.
Received12April2005;acceptedrevisedmanuscript13July2005.
NovelSequenceVariantsintheTMC1GeneinPakistaniFamilieswithAutosomalRecessiveHearingImpairmentRegieLynP.
Santos1,4,MuhammadWajid2,MohammadNasimKhan2,NathanMcArthur1,ThanhL.
Pham1,AttyaBhatti2,KwanghyukLee1,SabaIrshad2,AsifMir2,KaiYan1,MariaH.
Chahrour1,MuhammadAnsar3,WasimAhmad2andSuzanneM.
Leal1*1DepartmentofMolecularandHumanGenetics,BaylorCollegeofMedicine,Houston,Texas;2DepartmentofBiologicalSciences,Quaid-I-AzamUniversity,Islamabad,Pakistan;3DepartmentofBiosciences,COMSATSInstituteofInformationTechnology,Islamabad,Pakistan;4GeneticEpidemiologyUnit,DepartmentofEpidemiologyandBiostatistics,ErasmusMedicalCentre,Rotterdam,theNetherlands*Correspondenceto:Dr.
SuzanneM.
Leal,DepartmentofMolecularandHumanGenetics,BaylorCollegeofMedicine,OneBaylorPlaza,AlkekN1619.
01,Houston,Texas77030;Tel.
:1(713)798-4011;Fax:1(713)798-4373;E-mail:sleal@bcm.
tmc.
eduGrantsponsor:NationalInstitutesofHealth-NationalInstituteofDeafnessandOtherCommunicationDisorders(NIH-NIDCD)andHigherEducationCommission,Islamabad,Pakistan;Grantnumber:DC03594(NIH-NIDCD).
CommunicatedbyPaoloM.
FortinaThoughmanyhearingimpairmentgeneshavebeenidentified,onlyafewofthesegeneshavebeenscreenedinpopulationstudies.
Forthisstudy,168PakistanifamilieswithautosomalrecessivehearingimpairmentnotduetomutationsintheGJB2(Cx26)geneunderwentagenomescan.
Two-pointandmultipointparametriclinkageanalyseswerecarriedout.
Twelvefamilieshadtwo-pointormultipointLODscoresof1.
4orgreaterwithinthetransmembranecochlearexpressedgene1(TMC1)regionandweresubjectedtofurtherscreeningwithdirectDNAsequencing.
Fivenovelputativelyfunctionalnon-synonymoussequencevariants,c.
830A>G(p.
Y277C),c.
1114G>A(p.
V372M),c.
1334G>A(p.
R445H),c.
2004T>G(p.
S668R)andc.
2035G>A(p.
E679K),werefoundtosegregatewithinsevenfamilies,butwerenotobservedin234Pakistanicontrolchromosomes.
Thevariantsc.
830A>G(p.
Y277C),c.
1114G>A(p.
V372M)andc.
1334G>A(p.
R445H)occurredathighlyconservedregionsandwerepredictedtoliewithinhydrophobictransmembranedomains,whilenon-synonymousvariantsc.
2004T>G(p.
S668R)andc.
2035G>A(p.
E679K)occurredinextracellularregionsthatwerenothighlyconserved.
Thereisevidencethatthec.
2004T>G(p.
S668R)variantmayhaveoccurredataphosphorylationsite.
Onefamilyhastheknownsplicesitemutationc.
536-8T>A.
Theprevalenceofnon-syndromichearingimpairmentduetoTMC1inthisPakistanipopulationis4.
4%(95%CI:1.
9,8.
6%).
TheTMC1proteinmighthaveanimportantfunctioninK+channelsofinnerhaircells,whichwouldbeconsistentwiththehypotheticalstructureofproteindomainsinwhichsequencevariantswereidentified.
2005Wiley-Liss,Inc.
KEYWORDS:TMC1;autosomalrecessivenon-syndromichearingimpairment;Pakistan;prevalenceINTRODUCTIONDuetothehighlycomplexstructureandfunctionofthehumaninnerear,itisnotsurprisingthatsensorineuralhearingimpairment(HI)isgeneticallyheterogeneous.
Currently,37non-syndromichearingimpairment(NSHI)DOI:10.
1002/humu.
93742Santosetal.
geneshavebeenidentified,ofwhich21areassociatedwithautosomalrecessive(AR)NSHI(VanCampG,SmithRJH.
HereditaryHearingLossHomepage.
URL:http://webhost.
ua.
ac.
be/hhh/Accessed1March2005).
However,oftheARNSHIgenes,onlyafew[e.
g.
GJB2(MIM#121011),CDH23(MIM#605516)andWFS1(MIM#606201)]havebeenwellcharacterizedintermsofprevalenceandsequencevariantsfoundindifferentpopulations.
AdditionalstudiesofgenesthatareinvolvedinNSHIwithinvariouspopulationsarenecessarytobetterunderstandtheirprevalence,spectrumofsequencevariantsandpublichealthsignificance.
Inthisstudy,TMC1(MIM#606706)wasstudiedwithinthePakistanipopulation.
Atotalof168PakistanifamilieswithARNSHIthatwerenegativeforGJB2mutationsunderwenta10cMgenomescan.
Twelvefamilieshadtwo-pointormultipointLODscoresof>1.
4withinthe9q21.
13regionwhereTMC1isphysicallymapped.
ThesefamilieswereselectedforsequencingoftheTMC1gene.
ThereaftertheprevalenceofTMC1variantsinthispopulationwasestimated.
Also,bylocatingtheresiduesatpredictedtransmembranedomainsandstudyingevolutionaryconservationinmultiplesequencealignment,thepossibleeffectsontheTMC1proteinproductofbothnovelandpreviouslyreportedsequencevariantswereexamined.
MATERIALSANDMETHODSAscertainmentofstudysubjectsThestudywasapprovedbytheQuaid-I-AzamUniversityInstitutionalReviewBoardandbytheInstitutionalReviewBoardforHumanSubjectResearchforBaylorCollegeofMedicineandAffiliatedHospitals.
Informedconsentwasobtainedfromallfamilymemberswhoparticipatedinthestudy.
Forthisstudy,192unrelatedPakistanifamilieswithatleasttwoARNSHIindividualswereascertainedfromvariousregionsofPakistan.
Medicalandfamilyhistoryandinformationonpedigreestructurewasobtainedfrommultiplefamilymembers.
Puretoneaudiometryat250-8000Hzwasperformedforselectedsubjects.
Allhearing-impairedfamilymembersunderwentphysicalexamination.
Withinthesefamilies,noclinicalfeaturesthatwouldindicatethattheHIwaspartofasyndromewereobserved.
Inaddition,nogrossvestibularinvolvementwasnoted.
TheHIphenotypewasprelingual,severetoprofound,andwasnotknowntobecausedbyinflammatorymiddleeardiseaseorspecificenvironmentalfactors.
Additionally,117unrelatedhearingindividualswithoutafamilyhistoryofhearingimpairmentwereascertainedfromPakistan.
GenomescanDNAwasisolatedfromvenousbloodsamplesfollowingastandardprotocol(Grimbergetal.
,1989),quantifiedbyspectrophotometryatopticaldensity260,andstoredat-20°C.
Ofthe192families,twelvefamilieswerepositiveforGJB2(GenBankaccession#NM_004004.
3)mutations.
TwelveotherfamilieswerenotincludedforgenomescanduetoaninsufficientnumberofDNAsamplesinordertocarryoutalinkageanalysis.
DNAsamplesfrom126familiesweredilutedto40ηg/landsenttotheCenterforInheritedDiseaseResearch(CIDR;URL:http://www.
cidr.
jhmi.
edu/)forgenomescan,whiledilutedDNAsamplesfrom42familiesweresenttotheNationalHeart,LungandBloodInstitute(NHLBI)MammalianGenotypingService(CenterforMedicalGenetics,Marshfield,WI,USA;URL:http://research.
marshfieldclinic.
org/genetics/)forgenotyping.
From2002to2004,samplesweresentinsixbatches,withanaverageof395shorttandemrepeat(STR)markersspacedat~10cMapartforeachgenomescanthatwasdoneonall22autosomesandtheXandYchromosomes.
LinkageanalysisLinkageanalyseswereperformedunderafullypenetrantautosomalrecessivemodelwithadiseaseallelefrequencyof0.
001.
TheMLINKprogramoftheFASTLINKcomputerpackagewasutilizedfortwo-pointlinkageanalysis(Cottinghametal.
,1993),whilemultipointanalysiswasperformedusingALLEGRO(Gudbjartssonetal.
,2002)withmapdistancesfromtheMarshfieldgeneticmap(Bromanetal.
,1998).
SomeofthefamiliesweretoolargetoanalyzeintheirentiretyusingALLEGROandwerethereforebrokenintotwoormorebranchesfortheanalysis,thentheLODscoresfromeachbranchofthefamilyweresummed.
Markerallelefrequencieswereestimatedfromthedatabymeansofbothobservedandreconstructedgenotypesoffoundersfromeachpedigreeandtheotherpedigreesinthesamegenomescan.
TMC1VariantsinPakistaniFamilies3SequencingTMC1Polymerasechainreaction(PCR)primersweredesignedfor24exonsplus1000basepairsofthepromoterregionofTMC1(GenBankaccession#NM_138691.
2)viaPrimer3software(RozenandSkaletsky,2000).
DNAfromoneunaffectedandtwoaffectedindividualsfromeachfamilyweredilutedto5ηg/l,amplified,andpurifiedwithExoSAP-IT(USBCorp.
,Cleveland,Ohio,USA;URL:http://www.
usbweb.
com/).
SequencingwiththeappropriateprimerswasperformedwiththeBigDyeTerminatorv3.
1CycleSequencingKittogetherwithanAppliedBiosystems3700DNAAnalyzer(AppleraCorp.
,FosterCity,CA;URL:http://www.
appliedbiosystems.
com/).
SequencevariantswereidentifiedviaSequencherTMVersion4.
1.
4software(GeneCodesCorp.
,AnnArbor,MI,USA;URL:http://www.
genecodes.
com/sequencher/).
Whenasequencevariantwasfound,DNAsamplesfromtherestofthefamilyweresequencedfortheexoninwhichthevariantwasidentified.
Similarly,117controlindividualsfromPakistanwerescreenedforthesameexon.
ProteinsequenceanalysisTodeterminetheevolutionaryconservationofidentifiedsubstitutions,theExPASy(ExpertProteinAnalysisSystem;URL:http://us.
expasy.
org/)proteomicsserveroftheSwissInstituteofBioinformatics(SIB)wasusedtolookforhomologuesoftheTMC1protein.
ExPASyusestheNCBIBLASTP1.
5.
4-Paracelprogram(Altschuletal.
,1997)tosearchtheExPASy/UniProtdatabase.
ToperformtheBLASTPsearch,thedefaultsettingswereused,exceptthatthethresholdforexpectedrandommatches(E)wassettoamoreconservativevalueof10-4inordertominimizefalsepositiveresults.
Ofthe81querymatches(scorerange58-1196),onematchperspecieswaschosenforfurtheralignment.
Nineproteins,includingthehumanTMC1sequence,werethensubmittedformultiplesequencealignmentviaClustalW(Thompsonetal.
,1994)attheEuropeanBioinformaticsInstitute(EBI;URL:http://www.
ebi.
ac.
uk/clustalw/)usingdefaultsettings.
Membrane-spanningdomainsoftheTMC1proteinwerepredictedthroughtheTMHMMServerv.
2.
0(Kroghetal.
,2001).
AnindependentstudydemonstratedthatTMHMMv.
2.
0had92%truepositivepredictions,whichwerehigherthanallothertransmembraneproteinpredictionprograms(Molleretal.
,2001).
Tocheckformotifsorpatternswithintheproteinsequence,thePROSITEdatabaseofSIBandtheEuropeanMolecularBiologyLaboratory(EMBL)-EBIwasscanned(Sigristetal.
,2002).
ThewebserverPolyPhen,alsofromEMBL,wasusedtoassessthefunctionaleffectofidentifiedsubstitutions(Ramenskyetal.
,2002).
RESULTSOf168familiesthatunderwenttwo-pointand/ormultipointlinkageanalysis,twelvefamilieshadtwo-pointormultipointLODscoresof1.
4orgreaterbetweenmarkersflankingtheTMC1gene.
Table1showstheLODscoresperfamily,alongwithethno-linguisticinformationandsequencevariantsthatwerefoundtosegregatewithinfamilies.
Thesesequencevariantswerefoundinthehomozygousstateinthehearing-impairedindividualsbutnotamongunaffectedfamilymembers.
4Santosetal.
Table1.
FamiliesscreenedforTMC1mutations*FamilyPlaceoforiginLanguage2-pointLODMultipointLODSequencevariant4008DGKhan,PunjabSairiki3.
14.
0c.
2035G>A(p.
E679K)4027MianWali,NWFP-PunjabborderPunjabi2.
5;2.
6atchr.
111.
1;2.
0atchr.
11None4033Sarghoda,PunjabPunjabi2.
2;1.
2atchr.
22.
9;2.
1atchr.
2c.
1334G>A(p.
R445H)4049Bahwalpur,PunjabSairiki4.
74.
8c.
830A>G(p.
Y277C)4070Kashmir,AJKKashmiri2.
43.
0c.
2004T>G(p.
S668R)4090Chistian,PunjabPunjabi1.
4;2.
1atchr.
101.
8;-0.
6atchr.
10c.
536-8T>A4119Sadiqabad,PunjabSairiki2.
42.
6c.
1114G>A(p.
V372M)4138KotliKshmir,AJKKashmiri2.
63.
3c.
2004T>G(p.
S668R)4156Larkhana,SindSindi2.
2;2.
3atchr.
83.
1;0atchr.
8None4160Dhandi,SindSindi3.
03.
6c.
1114G>A(p.
V372M)4165TajGhar,PunjabSairiki1.
5;2.
0atchr.
172.
7;2.
8atchr.
17None4173Patoki,PunjabPunjabi1.
6;1.
7atchr.
11.
6;1.
9atchr.
1None*TMC1(GenBankaccession#NM_138691.
2).
NWFP=NorthwesternFrontierProvince;AJK=AzadJammunandKashmir.
Thetraditionalnomenclatureforc.
536-8T>AwasIVS10-8T>A.
Table2.
TMC1sequencevariantsfoundtosegregatewithinPakistanifamiliesExonNucleotidechangeAminoacidchangeDomain*EvolutionaryconservationPredictedfunctionaleffectAllelefrequenciesamongcontrolchromosomes11c.
536-8T>ASpliceacceptorN/AN/AExon11skipped0/23413c.
830A>Gp.
Y277CTM2IdenticalProbablydamaging0/23415c.
1114G>Ap.
V372MTM3ConservedBenign0/23416c.
1334G>Ap.
R445HTM4IdenticalBenign0/23421c.
2004T>Gp.
S668RECNon-conservedBenign;PKCphosphorylationsite§0/23421c.
2035G>Ap.
E679KECNon-conservedBenign0/234*TMHMMv.
2.
0predictedsixmembrane-spanningdomainsfortheTMC1protein(Kroghetal.
,2001).
TM=transmembrane;EC=extracellular;N/A=notapplicable.
HumanTMC1sequencecomparedwithTMC1-likeproteinsequencesof8specieswhichwereidentifiedviatheNCBIBLASTP(Altschuletal.
,1997)programandtheExPASy/UniProtdatabases.
Speciesinclude1mammal,1amphibian,2bonyfishes,3insectsand1nematode.
ComparisondoneviaClustalW(Thompsonetal.
,1994).
Theserineresidueatposition668andglutamicacidat679areconservedinM.
musculusTmc1andF.
rubripesTmc2-relatedproteins.
Additionally,S668isalsoconservedinA.
gambiaeTmcA-likeproteinandD.
melanogasterCG3280-PA.
PossibleeffectofaminoacidsubstitutionsontheproteinstructureandfunctionwerepredictedviathewebserverPolyPhen(Ramenskyetal.
,2002).
§Thep.
S668RsequencechangewasfoundtooccuratapossibleproteinkinaseC(PKC)phosphorylationsiteaccordingtothePROSITEdatabase(Sigristetal.
,2002).
TMC1VariantsinPakistaniFamilies5Eightfamilieswerepositiveforputativelyfunctionalsequencevariants.
Oftheidentifiedvariants,c.
830A>G(p.
Y277C),whichoccurredatahighlyconservedresidue,wasdeemedtobedamagingbecauseofachangefromahydroxyltoasulfhydrylsidechainintheaminoacidthatwaslocatedwithinthesecondtransmembrane(TM)domain(Table2).
Thec.
1114G>A(p.
V372M)andc.
1334G>A(p.
R445H)variantswerealsoathighlyconservedTMresidues,buttheaminoacidwaschangedtoanotherofthesamesubclass.
Residuesforvariantsc.
2004T>G(p.
S668R)andc.
2035G>A(p.
E679K)werelessconservedandoccurredwithintheextracellularregion.
TheselastfoursequencechangeswereconsideredbenignbythePolyphenwebsite,thoughscanningthroughPROSITEshowedthattheserineresidueatposition668maybelongtoaproteinkinaseCphosphorylationsite.
Thenucleotidechangec.
2004T>Cremovesthisputativephosphorylationsite.
Alsoc.
1114G>A(p.
V372M)andc.
2004T>G(p.
S668R)wereeachfoundtosegregatewiththeNSHIstatusintwofamilies.
Onefamilyhastheknownsplicesitemutationc.
536-8T>A(Kurimaetal.
,2002).
Allthereportedsequencevariantswerenotfoundincontrolsubjects,whowerelinguisticallymatchedforthePunjabi-andSairiki-speakingfamilies.
DuetothelimitedsamplesfromtheremoteSindiandKashmiriregions,mostofthecontrolsforthec.
2004T>GvariantarePunjabi,whileforthec.
1114G>AvariantmostofthecontrolsspeakSairiki.
Severalpolymorphismswerealsoobservedinthetwelvefamilies(Table3).
Noneofthesevariantssegregatedwithhearingimpairmentstatuswithinthefamilies.
Twoofthevariantswerenon-synonymoussubstitutions,butwerealsoconsideredbenignpolymorphismsbecausetheydidnotsegregatewithHIstatus.
Thesesubstitutionsalsooccurredatnon-conservednon-transmembraneresidues.
Asidefromc.
536-8T>A,otherTMC1sequencevariantsthatwereidentifiedinpreviousresearchwerenotobservedinthefamiliesinthisstudy,butwerealsoanalyzedintermsofevolutionaryconservationandoccurrenceattransmembranedomainsandknownmotifs(Table4).
Table3.
TMC1polymorphismsthatdonotsegregatewithinfamiliesExonNucleotideChange1g.
74G>A(-467fromtheATG)3g.
[236-67G>A(+)322A>G(-219fromtheATG)]4g.
346-11delT6c.
45C>T(synonymous)8c.
237-44G>A8c.
241G>A(p.
E81K)*10c.
[535+101A>G(+)535+106_113delCAAACAAA]14c.
1029+85T>C16c.
1404+32A>G17c.
1457T>C(p.
M486T)22c.
2208+22A>G*p.
E81KdoesnotsegregatewiththeHIstatusinthreefamilies.
Additionallyp.
E81Kisattheintracellularglutamicacid(E)-richregionandisnon-conserved.
p.
M486Twasfoundintheheterozygousstateintwohearingindividualsfromtwodifferentfamiliesbutnotamongthehearing-impaired.
Theresidueispredictedtobeintracellularandisnon-conserved.
6Santosetal.
Table4.
PreviouslypublishedsequencevariantsintheTMC1gene*ExonNucleotideChangeProteinChangeDomainEvolutionaryConservationPredictedFunctionalEffectIntron3-5IVS3_IVS5del27kbTranscriptioninitiationsiteremovedN/AN/AExons4and5deleted7c.
100C>Tp.
R34XICNon-conservedTruncatedprotein;Occursatglutamic-acidrichN-terminusregion;cAMP-andcGMP-dependentproteinkinasephosphorylationsite8c.
295delAFrameshiftN/AN/ATruncatedprotein13c.
884+1G>ASplicedonorN/AN/AExon13skipped15c.
1165C>Tp.
R389XECNon-conservedTruncatedprotein17c.
1534C>Tp.
R512XICNon-conservedTruncatedprotein19c.
1714G>A§p.
D572NICNon-conservedBenign;CaseinkinaseIIphosphorylationsite20c.
1960A>Gp.
M654VTM5Non-conservedBenign*TMC1sequencechangeswerereportedbyKurimaetal.
(2002)exceptforp.
R389Xatexon15(Meyeretal.
,2005).
Membrane-spanningdomains,evolutionaryconservationandfunctionaleffectweredeterminedasdescribedinTable2.
Becausethearticledidnotspecifytheexactpositionatwhichthisdeletionoccurred,itwasnotpossibletorenamethissequencevariantaccordingtothecurrentnomenclaturerecommendations.
Thetraditionalnomenclatureforc.
884+1G>AwasIVS13+1G>A.
§c.
1714G>A(p.
D572N)wasobservedinaNorthAmericanfamilywithdominanthearingimpairment.
TMC1VariantsinPakistaniFamilies7DISCUSSIONPutativelyfunctionalTMC1variantswereobservedineightofthe168Pakistanifamiliesthatunderwentagenomescan.
TwelveadditionalfamiliesinthestudydidnotundergoagenomescanbutareknowntohavefunctionalvariantsintheGJB2gene(Santosetal.
,2005).
Thus,whenGJB2-affectedfamilieswereincluded,theprevalenceofTMC1mutationsinthisPakistanipopulationwas4.
4%(95%CI:1.
9,8.
6).
Thisisalowerprevalenceratethanthepreviouslyreportedprevalenceof5.
4+3.
0%amongconsanguineousIndianandPakistanifamilies(Kurimaetal.
,2002).
However,thedifferenceintheseprevalenceratesisnotstatisticallysignificant.
Among12familiesinwhichtheTMC1genewassequenced,apotentiallyfunctionalvariantwasnotidentifiedinfourfamilies(Table1).
Threeofthesefamilies(4027,4165and4173)hadLODscoresofsuggestivelinkage(1.
9-2.
8)inotherchromosomalregions,whichmaycontainthegenethatcausestheHIinthesefamilies.
Infamily4156,thetwo-pointLODscorewasrelativelyhighforchromosome2,butthemultipointLODscoreatchromosome2waszero,whileamultipointLODscoreof3.
1wasobtainedintheTMC1region.
Forthisfamily,itispossiblethatthefunctionalvariant(s)arewithintheintronicregionsofTMC1,orinanothergenethatliesclosetotheTMC1gene.
ThepredictedstructureofTMC1bearssimilaritytothatoftheα-subunitofvoltage-dependentK+channels,whichhassixα-helicalTMsegmentsandintracellularNandCtermini(HanlonandWallace,2002).
ThefirstfourTMdomainsoftheK+channelα-subunitactasvoltagesensorsforactivationgating(Li-Smerinetal.
,2000),whereastheinterveningsegmentbetweenTM5andTM6appearstoconferchannelselectivity(HanlonandWallace,2002).
ThreehighlyconservedTMC1sequencevariantsinthisstudy–c.
830A>G(p.
Y277C),c.
1114G>A(p.
V372M)andc.
1334G>A(p.
R445H)–liewithinthecentralportionofhydrophobicTMsegments.
Meanwhile,theTMC1variantsc.
2004T>G(p.
S668R)andc.
2035G>A(p.
E679K)werefoundbetweenTM5andTM6amongaclusteroflessconservedresidues.
Thoughtheseaminoacidresidueswerenon-conservedwhenhumanTMC1wasalignedwitheightproteinsfromotherspecies,alignmentwithotherhumanandmurineTMCproteinsshowsthattheseresiduesareconservedformembersoftheTMCsubfamilyA,TMC1,TMC2andTMC3(Keresztesetal.
,2003;Kurimaetal.
,2003).
Additionally,theserineresidueatposition668isaputativeproteinkinaseCphosphorylationsite.
Therefore,eventhoughthePolyphenserverlabeledfouroffivenovelsubstitutionsasbenign,webelievethatallfivevariantsarefunctional.
ItshouldbenotedthatthePolyphenserversuccessfullypredictsonly82%ofmutationsintheSwissProtproteindatabase(Ramenskyetal.
,2002).
Thedeafness(dn)mouseisthehomologousmodelforrecessiveTMC1mutations,whiletheBeethoven(Bth)mouseisidentifiedwithdominantTMC1mutationsthatcausepostlingualhearingimpairment(Kurimaetal.
,2002;Vreugdeetal.
,2002).
InthemouseTmc1expressionhasbeenfoundtolocalizetocochlearhaircells(Kurimaetal.
,2002).
Inmicroscopicsectionsofdnmousecochlea,Tmc1mutationscausedthegreatestdamagetoinnerhaircells(IHC),withprogressivedegenerationthatbeginsatpostnatalday13,whichisthetimeofonsetofhearingfunctioninmice(Vreugdeetal.
,2002;Pujoletal.
,1983).
Infunctionalstudies,thecochlearmicrophonic,whichisameasureoftheelectricpotentialthatisgeneratedbytheorganofCortiafteracousticstimulation,wasneverdetectedindnmicefrompostnatalday12onwards(SteelandBock,1980;BockandSteel,1983).
ThesestudiesfurthersupportthefindingsofbilateralprofoundprelingualhearingimpairmentduetorecessivehomozygousTMC1mutations,andimplicatetheIHCasthemainsiteofhearingdysfunction.
ItwassuggestedthatTMC1mightbeanionchannelortransporterwhichmediatesK+homeostasisintheinnerear(Keresztesetal.
,2003).
BecauseK+transportacrosshaircellmembranesrequiresveryrapidresponse,theuseofsecondmessengerorcascademechanismshasbeendeemedhighlyunlikely,andthegating-springmodelformechanoelectricaltransductionislargelyfavored(CoreyandHudspeth,1983).
Thismodelproposedthatmechanicalforcesbroughtaboutbybendingofstereociliaandtensiononthetiplinksdirectlyactivateionchannels.
IfitistruethatTMC1isanionchannelthatismainlylocalizedintheIHC,thenitmightbeinvolvedinthemostbasicauditoryprocessofhair-celltransduction.
Inconclusion,fivenovelsequencevariantsoftheTMC1geneforhearingimpairmentwereidentifiedinthisstudy.
ThesevariantswerefoundinTMC1regionsthatmightbecriticaltofunctionofK+channelsinhaircells.
InthehighlyconsanguineousPakistanipopulation,theprevalenceofputativelyfunctionalTMC1variantswasfoundtobelow.
However,thetrueimpactofgeneticdefectsinTMC1cannotbeknownuntilmorestudiesaredoneinotherpopulations.
ACKNOWLEDGMENTSWewishtothankthefamilymembersfortheirinvaluableparticipationandcooperation.
WearealsogratefultoRAHarrisforhisusefulcommentsandsuggestions.
ThisworkwasmadepossiblethroughtheNationalInstitutes8Santosetal.
ofHealth-NationalInstituteofDeafnessandOtherCommunicationDisordersgrantR01-DC03594.
GenotypingserviceswereprovidedbytheCenterforInheritedDiseaseResearch(CIDR)andtheNationalHeart,LungandBloodInstituteMammalianGenotypingService.
CIDRisfullyfundedthroughafederalcontractfromtheNationalInstitutesofHealthtoTheJohnsHopkinsUniversity,ContractNumberN01-HG-65403.
REFERENCESAltschulSF,MaddenTL,SchafferAA,ZhangJ,ZhangZ,MillerW,LipmanDJ.
1997.
GappedBLASTandPSI-BLAST:anewgenerationofproteindatabasesearchprograms.
NucleicAcidsRes25:3389-3402.
BockGR,SteelKP.
1983.
Innerearpathologyinthedeafnessmutantmouse.
ActaOtolaryngol96:39-47.
BromanKW,MurrayJC,SheffieldVC,WhiteRL,WeberJL.
1998.
Comprehensivehumangeneticmaps:individualandsex-specificvariationinrecombination.
AmJHumGenet63:861-869.
CoreyDP,HudspethAJ.
1983.
Kineticsofthereceptorcurrentinbullfrogsaccularhaircells.
JNeurosci3:962-976.
CottinghamRW,IduryRM,SchafferAA.
1993.
Fastersequentialgeneticlinkagecomputations.
AmJHumGenet.
53:252-263.
GrimbergJ,NawoschikS,BellusicoL,McKeeR,TurckA,EisenbergA.
1989.
Asimpleandefficientnon-organicprocedurefortheisolationofgenomicDNAfromblood.
NucleicAcidRes17:83-90.
GudbjartssonDF,JonassonK,FriggeML,KongA.
2000.
Allegro,anewcomputerprogramformultipointlinkageanalysis.
NatGenet25:12-13.
HanlonMR,WallaceBA.
2002.
Structureandfunctionofvoltage-dependentionchannelregulatorybetasubunits.
Biochemistry41:2886-2894.
KeresztesG,MutaiH,HellerS.
2003.
TMCandEVERgenesbelongtoalargernovelfamily,theTMCgenefamilyencodingtransmembraneproteins.
BMCGenomics4:24.
KroghA,LarssonB,vonHeijneG,SonnhammerEL.
2001.
PredictingtransmembraneproteintopologywithahiddenMarkovmodel:Applicationtocompletegenomes.
JMolBiol305:567-580.
KurimaK,PetersLM,YangY,RiazuddinS,AhmedZM,NazS,ArnaudD,DruryS,MoJ,MakishimaT,GhoshM,MenonPS,DeshmukhD,OddouxC,OstrerH,KhanS,RiazuddinS,DeiningerPL,HamptonLL,SullivanSL,BatteyJFJr,KeatsBJ,WilcoxER,FriedmanTB,GriffithAJ.
2002.
Dominantandrecessivedeafnesscausedbymutationsofanovelgene,TMC1,requiredforcochlearhair-cellfunction.
NatGenet30:277-284.
KurimaK,YangY,SorberK,GriffithAJ.
2003.
Characterizationofthetransmembranechannel-like(TMC)genefamily:functionalcluesfromhearinglossandepidermodysplasiaverruciformis.
Genomics82:300-308.
Li-SmerinY,HackosDH,SwartzKJ.
2000.
α-helicalstructuralelementswithinthevoltage-sensingdomainsofaK(+)channel.
JGenPhysiol115:33-50.
MeyerCG,GasmelseedNM,MerganiA,MagzoubMMA,MuntauB,ThorstenT,HorstmannRD.
2005.
NovelTMC1structuralandsplicevariantsassociatedwithcongenitalnonsyndromicdeafnessinaSudanesepedigree.
HumMutat25:100.
MollerS,CroningMD,ApweilerR.
2001.
Evaluationofmethodsforthepredictionofmembranespanningregions.
Bioinformatics17:646-653.
PujolR,ShnersonA,LenoirM,DeolMS.
1983.
Earlydegenerationofsensoryandganglioncellsintheinnerearofmicewithuncomplicatedgeneticdeafness(dn):preliminaryobservations.
HearRes12:57-63.
RamenskyV,BorkP,SunyaevS.
2002.
Humannon-synonymousSNPs:serverandsurvey.
NucleicAcidsRes30:3894-3900.
RozenS,SkaletskyHJ.
2000.
Primer3ontheWWWforgeneralusersandforbiologistprogrammers.
In:KrawetzS,MisenerS,editors.
BioinformaticsMethodsandProtocols:MethodsinMolecularBiology.
Totowa,NJ:HumanaPress.
p365-386.
SantosRLP,WajidM,PhamTL,HussanJ,AliG,AhmadW,LealSM.
2005.
LowprevalenceofConnexin26(GJB2)variantsinPakistanifamilieswithautosomalrecessivenon-syndromichearingimpairment.
ClinGenet67:61-68.
TMC1VariantsinPakistaniFamilies9SigristCJ,CeruttiL,HuloN,GattikerA,FalquetL,PagniM,BairochA,BucherP.
2002.
PROSITE:adocumenteddatabaseusingpatternsandprofilesasmotifdescriptors.
BriefBioinform3:265-274.
SteelKP,BockGR.
1980.
Thenatureofinheriteddeafnessindeafnessmice.
Nature288:159-161.
ThompsonJD,HigginsDG,GibsonTJ.
1994.
CLUSTALW:improvingthesensitivityofprogressivemultiplesequencealignmentthroughsequenceweighting,position-specificgappenaltiesandweightmatrixchoice.
NucleicAcidsRes22:4673-4680.
VreugdeS,ErvenA,KrosCJ,MarcottiW,FuchsH,KurimaK,WilcoxER,FriedmanTB,GriffithAJ,BallingR,HrabeDeAngelisM,AvrahamKB,SteelKP.
2002.
Beethoven,amousemodelfordominant,progressivehearinglossDFNA36.
NatGenet30:257-258.
Received12April2005;acceptedrevisedmanuscript13July2005.
NovelSequenceVariantsintheTMC1GeneinPakistaniFamilieswithAutosomalRecessiveHearingImpairmentRegieLynP.
Santos1,4,MuhammadWajid2,MohammadNasimKhan2,NathanMcArthur1,ThanhL.
Pham1,AttyaBhatti2,KwanghyukLee1,SabaIrshad2,AsifMir2,KaiYan1,MariaH.
Chahrour1,MuhammadAnsar3,WasimAhmad2andSuzanneM.
Leal1*1DepartmentofMolecularandHumanGenetics,BaylorCollegeofMedicine,Houston,Texas;2DepartmentofBiologicalSciences,Quaid-I-AzamUniversity,Islamabad,Pakistan;3DepartmentofBiosciences,COMSATSInstituteofInformationTechnology,Islamabad,Pakistan;4GeneticEpidemiologyUnit,DepartmentofEpidemiologyandBiostatistics,ErasmusMedicalCentre,Rotterdam,theNetherlands*Correspondenceto:Dr.
SuzanneM.
Leal,DepartmentofMolecularandHumanGenetics,BaylorCollegeofMedicine,OneBaylorPlaza,AlkekN1619.
01,Houston,Texas77030;Tel.
:1(713)798-4011;Fax:1(713)798-4373;E-mail:sleal@bcm.
tmc.
eduGrantsponsor:NationalInstitutesofHealth-NationalInstituteofDeafnessandOtherCommunicationDisorders(NIH-NIDCD)andHigherEducationCommission,Islamabad,Pakistan;Grantnumber:DC03594(NIH-NIDCD).
CommunicatedbyPaoloM.
FortinaThoughmanyhearingimpairmentgeneshavebeenidentified,onlyafewofthesegeneshavebeenscreenedinpopulationstudies.
Forthisstudy,168PakistanifamilieswithautosomalrecessivehearingimpairmentnotduetomutationsintheGJB2(Cx26)geneunderwentagenomescan.
Two-pointandmultipointparametriclinkageanalyseswerecarriedout.
Twelvefamilieshadtwo-pointormultipointLODscoresof1.
4orgreaterwithinthetransmembranecochlearexpressedgene1(TMC1)regionandweresubjectedtofurtherscreeningwithdirectDNAsequencing.
Fivenovelputativelyfunctionalnon-synonymoussequencevariants,c.
830A>G(p.
Y277C),c.
1114G>A(p.
V372M),c.
1334G>A(p.
R445H),c.
2004T>G(p.
S668R)andc.
2035G>A(p.
E679K),werefoundtosegregatewithinsevenfamilies,butwerenotobservedin234Pakistanicontrolchromosomes.
Thevariantsc.
830A>G(p.
Y277C),c.
1114G>A(p.
V372M)andc.
1334G>A(p.
R445H)occurredathighlyconservedregionsandwerepredictedtoliewithinhydrophobictransmembranedomains,whilenon-synonymousvariantsc.
2004T>G(p.
S668R)andc.
2035G>A(p.
E679K)occurredinextracellularregionsthatwerenothighlyconserved.
Thereisevidencethatthec.
2004T>G(p.
S668R)variantmayhaveoccurredataphosphorylationsite.
Onefamilyhastheknownsplicesitemutationc.
536-8T>A.
Theprevalenceofnon-syndromichearingimpairmentduetoTMC1inthisPakistanipopulationis4.
4%(95%CI:1.
9,8.
6%).
TheTMC1proteinmighthaveanimportantfunctioninK+channelsofinnerhaircells,whichwouldbeconsistentwiththehypotheticalstructureofproteindomainsinwhichsequencevariantswereidentified.
2005Wiley-Liss,Inc.
KEYWORDS:TMC1;autosomalrecessivenon-syndromichearingimpairment;Pakistan;prevalenceINTRODUCTIONDuetothehighlycomplexstructureandfunctionofthehumaninnerear,itisnotsurprisingthatsensorineuralhearingimpairment(HI)isgeneticallyheterogeneous.
Currently,37non-syndromichearingimpairment(NSHI)DOI:10.
1002/humu.
93742Santosetal.
geneshavebeenidentified,ofwhich21areassociatedwithautosomalrecessive(AR)NSHI(VanCampG,SmithRJH.
HereditaryHearingLossHomepage.
URL:http://webhost.
ua.
ac.
be/hhh/Accessed1March2005).
However,oftheARNSHIgenes,onlyafew[e.
g.
GJB2(MIM#121011),CDH23(MIM#605516)andWFS1(MIM#606201)]havebeenwellcharacterizedintermsofprevalenceandsequencevariantsfoundindifferentpopulations.
AdditionalstudiesofgenesthatareinvolvedinNSHIwithinvariouspopulationsarenecessarytobetterunderstandtheirprevalence,spectrumofsequencevariantsandpublichealthsignificance.
Inthisstudy,TMC1(MIM#606706)wasstudiedwithinthePakistanipopulation.
Atotalof168PakistanifamilieswithARNSHIthatwerenegativeforGJB2mutationsunderwenta10cMgenomescan.
Twelvefamilieshadtwo-pointormultipointLODscoresof>1.
4withinthe9q21.
13regionwhereTMC1isphysicallymapped.
ThesefamilieswereselectedforsequencingoftheTMC1gene.
ThereaftertheprevalenceofTMC1variantsinthispopulationwasestimated.
Also,bylocatingtheresiduesatpredictedtransmembranedomainsandstudyingevolutionaryconservationinmultiplesequencealignment,thepossibleeffectsontheTMC1proteinproductofbothnovelandpreviouslyreportedsequencevariantswereexamined.
MATERIALSANDMETHODSAscertainmentofstudysubjectsThestudywasapprovedbytheQuaid-I-AzamUniversityInstitutionalReviewBoardandbytheInstitutionalReviewBoardforHumanSubjectResearchforBaylorCollegeofMedicineandAffiliatedHospitals.
Informedconsentwasobtainedfromallfamilymemberswhoparticipatedinthestudy.
Forthisstudy,192unrelatedPakistanifamilieswithatleasttwoARNSHIindividualswereascertainedfromvariousregionsofPakistan.
Medicalandfamilyhistoryandinformationonpedigreestructurewasobtainedfrommultiplefamilymembers.
Puretoneaudiometryat250-8000Hzwasperformedforselectedsubjects.
Allhearing-impairedfamilymembersunderwentphysicalexamination.
Withinthesefamilies,noclinicalfeaturesthatwouldindicatethattheHIwaspartofasyndromewereobserved.
Inaddition,nogrossvestibularinvolvementwasnoted.
TheHIphenotypewasprelingual,severetoprofound,andwasnotknowntobecausedbyinflammatorymiddleeardiseaseorspecificenvironmentalfactors.
Additionally,117unrelatedhearingindividualswithoutafamilyhistoryofhearingimpairmentwereascertainedfromPakistan.
GenomescanDNAwasisolatedfromvenousbloodsamplesfollowingastandardprotocol(Grimbergetal.
,1989),quantifiedbyspectrophotometryatopticaldensity260,andstoredat-20°C.
Ofthe192families,twelvefamilieswerepositiveforGJB2(GenBankaccession#NM_004004.
3)mutations.
TwelveotherfamilieswerenotincludedforgenomescanduetoaninsufficientnumberofDNAsamplesinordertocarryoutalinkageanalysis.
DNAsamplesfrom126familiesweredilutedto40ηg/landsenttotheCenterforInheritedDiseaseResearch(CIDR;URL:http://www.
cidr.
jhmi.
edu/)forgenomescan,whiledilutedDNAsamplesfrom42familiesweresenttotheNationalHeart,LungandBloodInstitute(NHLBI)MammalianGenotypingService(CenterforMedicalGenetics,Marshfield,WI,USA;URL:http://research.
marshfieldclinic.
org/genetics/)forgenotyping.
From2002to2004,samplesweresentinsixbatches,withanaverageof395shorttandemrepeat(STR)markersspacedat~10cMapartforeachgenomescanthatwasdoneonall22autosomesandtheXandYchromosomes.
LinkageanalysisLinkageanalyseswereperformedunderafullypenetrantautosomalrecessivemodelwithadiseaseallelefrequencyof0.
001.
TheMLINKprogramoftheFASTLINKcomputerpackagewasutilizedfortwo-pointlinkageanalysis(Cottinghametal.
,1993),whilemultipointanalysiswasperformedusingALLEGRO(Gudbjartssonetal.
,2002)withmapdistancesfromtheMarshfieldgeneticmap(Bromanetal.
,1998).
SomeofthefamiliesweretoolargetoanalyzeintheirentiretyusingALLEGROandwerethereforebrokenintotwoormorebranchesfortheanalysis,thentheLODscoresfromeachbranchofthefamilyweresummed.
Markerallelefrequencieswereestimatedfromthedatabymeansofbothobservedandreconstructedgenotypesoffoundersfromeachpedigreeandtheotherpedigreesinthesamegenomescan.
TMC1VariantsinPakistaniFamilies3SequencingTMC1Polymerasechainreaction(PCR)primersweredesignedfor24exonsplus1000basepairsofthepromoterregionofTMC1(GenBankaccession#NM_138691.
2)viaPrimer3software(RozenandSkaletsky,2000).
DNAfromoneunaffectedandtwoaffectedindividualsfromeachfamilyweredilutedto5ηg/l,amplified,andpurifiedwithExoSAP-IT(USBCorp.
,Cleveland,Ohio,USA;URL:http://www.
usbweb.
com/).
SequencingwiththeappropriateprimerswasperformedwiththeBigDyeTerminatorv3.
1CycleSequencingKittogetherwithanAppliedBiosystems3700DNAAnalyzer(AppleraCorp.
,FosterCity,CA;URL:http://www.
appliedbiosystems.
com/).
SequencevariantswereidentifiedviaSequencherTMVersion4.
1.
4software(GeneCodesCorp.
,AnnArbor,MI,USA;URL:http://www.
genecodes.
com/sequencher/).
Whenasequencevariantwasfound,DNAsamplesfromtherestofthefamilyweresequencedfortheexoninwhichthevariantwasidentified.
Similarly,117controlindividualsfromPakistanwerescreenedforthesameexon.
ProteinsequenceanalysisTodeterminetheevolutionaryconservationofidentifiedsubstitutions,theExPASy(ExpertProteinAnalysisSystem;URL:http://us.
expasy.
org/)proteomicsserveroftheSwissInstituteofBioinformatics(SIB)wasusedtolookforhomologuesoftheTMC1protein.
ExPASyusestheNCBIBLASTP1.
5.
4-Paracelprogram(Altschuletal.
,1997)tosearchtheExPASy/UniProtdatabase.
ToperformtheBLASTPsearch,thedefaultsettingswereused,exceptthatthethresholdforexpectedrandommatches(E)wassettoamoreconservativevalueof10-4inordertominimizefalsepositiveresults.
Ofthe81querymatches(scorerange58-1196),onematchperspecieswaschosenforfurtheralignment.
Nineproteins,includingthehumanTMC1sequence,werethensubmittedformultiplesequencealignmentviaClustalW(Thompsonetal.
,1994)attheEuropeanBioinformaticsInstitute(EBI;URL:http://www.
ebi.
ac.
uk/clustalw/)usingdefaultsettings.
Membrane-spanningdomainsoftheTMC1proteinwerepredictedthroughtheTMHMMServerv.
2.
0(Kroghetal.
,2001).
AnindependentstudydemonstratedthatTMHMMv.
2.
0had92%truepositivepredictions,whichwerehigherthanallothertransmembraneproteinpredictionprograms(Molleretal.
,2001).
Tocheckformotifsorpatternswithintheproteinsequence,thePROSITEdatabaseofSIBandtheEuropeanMolecularBiologyLaboratory(EMBL)-EBIwasscanned(Sigristetal.
,2002).
ThewebserverPolyPhen,alsofromEMBL,wasusedtoassessthefunctionaleffectofidentifiedsubstitutions(Ramenskyetal.
,2002).
RESULTSOf168familiesthatunderwenttwo-pointand/ormultipointlinkageanalysis,twelvefamilieshadtwo-pointormultipointLODscoresof1.
4orgreaterbetweenmarkersflankingtheTMC1gene.
Table1showstheLODscoresperfamily,alongwithethno-linguisticinformationandsequencevariantsthatwerefoundtosegregatewithinfamilies.
Thesesequencevariantswerefoundinthehomozygousstateinthehearing-impairedindividualsbutnotamongunaffectedfamilymembers.
4Santosetal.
Table1.
FamiliesscreenedforTMC1mutations*FamilyPlaceoforiginLanguage2-pointLODMultipointLODSequencevariant4008DGKhan,PunjabSairiki3.
14.
0c.
2035G>A(p.
E679K)4027MianWali,NWFP-PunjabborderPunjabi2.
5;2.
6atchr.
111.
1;2.
0atchr.
11None4033Sarghoda,PunjabPunjabi2.
2;1.
2atchr.
22.
9;2.
1atchr.
2c.
1334G>A(p.
R445H)4049Bahwalpur,PunjabSairiki4.
74.
8c.
830A>G(p.
Y277C)4070Kashmir,AJKKashmiri2.
43.
0c.
2004T>G(p.
S668R)4090Chistian,PunjabPunjabi1.
4;2.
1atchr.
101.
8;-0.
6atchr.
10c.
536-8T>A4119Sadiqabad,PunjabSairiki2.
42.
6c.
1114G>A(p.
V372M)4138KotliKshmir,AJKKashmiri2.
63.
3c.
2004T>G(p.
S668R)4156Larkhana,SindSindi2.
2;2.
3atchr.
83.
1;0atchr.
8None4160Dhandi,SindSindi3.
03.
6c.
1114G>A(p.
V372M)4165TajGhar,PunjabSairiki1.
5;2.
0atchr.
172.
7;2.
8atchr.
17None4173Patoki,PunjabPunjabi1.
6;1.
7atchr.
11.
6;1.
9atchr.
1None*TMC1(GenBankaccession#NM_138691.
2).
NWFP=NorthwesternFrontierProvince;AJK=AzadJammunandKashmir.
Thetraditionalnomenclatureforc.
536-8T>AwasIVS10-8T>A.
Table2.
TMC1sequencevariantsfoundtosegregatewithinPakistanifamiliesExonNucleotidechangeAminoacidchangeDomain*EvolutionaryconservationPredictedfunctionaleffectAllelefrequenciesamongcontrolchromosomes11c.
536-8T>ASpliceacceptorN/AN/AExon11skipped0/23413c.
830A>Gp.
Y277CTM2IdenticalProbablydamaging0/23415c.
1114G>Ap.
V372MTM3ConservedBenign0/23416c.
1334G>Ap.
R445HTM4IdenticalBenign0/23421c.
2004T>Gp.
S668RECNon-conservedBenign;PKCphosphorylationsite§0/23421c.
2035G>Ap.
E679KECNon-conservedBenign0/234*TMHMMv.
2.
0predictedsixmembrane-spanningdomainsfortheTMC1protein(Kroghetal.
,2001).
TM=transmembrane;EC=extracellular;N/A=notapplicable.
HumanTMC1sequencecomparedwithTMC1-likeproteinsequencesof8specieswhichwereidentifiedviatheNCBIBLASTP(Altschuletal.
,1997)programandtheExPASy/UniProtdatabases.
Speciesinclude1mammal,1amphibian,2bonyfishes,3insectsand1nematode.
ComparisondoneviaClustalW(Thompsonetal.
,1994).
Theserineresidueatposition668andglutamicacidat679areconservedinM.
musculusTmc1andF.
rubripesTmc2-relatedproteins.
Additionally,S668isalsoconservedinA.
gambiaeTmcA-likeproteinandD.
melanogasterCG3280-PA.
PossibleeffectofaminoacidsubstitutionsontheproteinstructureandfunctionwerepredictedviathewebserverPolyPhen(Ramenskyetal.
,2002).
§Thep.
S668RsequencechangewasfoundtooccuratapossibleproteinkinaseC(PKC)phosphorylationsiteaccordingtothePROSITEdatabase(Sigristetal.
,2002).
TMC1VariantsinPakistaniFamilies5Eightfamilieswerepositiveforputativelyfunctionalsequencevariants.
Oftheidentifiedvariants,c.
830A>G(p.
Y277C),whichoccurredatahighlyconservedresidue,wasdeemedtobedamagingbecauseofachangefromahydroxyltoasulfhydrylsidechainintheaminoacidthatwaslocatedwithinthesecondtransmembrane(TM)domain(Table2).
Thec.
1114G>A(p.
V372M)andc.
1334G>A(p.
R445H)variantswerealsoathighlyconservedTMresidues,buttheaminoacidwaschangedtoanotherofthesamesubclass.
Residuesforvariantsc.
2004T>G(p.
S668R)andc.
2035G>A(p.
E679K)werelessconservedandoccurredwithintheextracellularregion.
TheselastfoursequencechangeswereconsideredbenignbythePolyphenwebsite,thoughscanningthroughPROSITEshowedthattheserineresidueatposition668maybelongtoaproteinkinaseCphosphorylationsite.
Thenucleotidechangec.
2004T>Cremovesthisputativephosphorylationsite.
Alsoc.
1114G>A(p.
V372M)andc.
2004T>G(p.
S668R)wereeachfoundtosegregatewiththeNSHIstatusintwofamilies.
Onefamilyhastheknownsplicesitemutationc.
536-8T>A(Kurimaetal.
,2002).
Allthereportedsequencevariantswerenotfoundincontrolsubjects,whowerelinguisticallymatchedforthePunjabi-andSairiki-speakingfamilies.
DuetothelimitedsamplesfromtheremoteSindiandKashmiriregions,mostofthecontrolsforthec.
2004T>GvariantarePunjabi,whileforthec.
1114G>AvariantmostofthecontrolsspeakSairiki.
Severalpolymorphismswerealsoobservedinthetwelvefamilies(Table3).
Noneofthesevariantssegregatedwithhearingimpairmentstatuswithinthefamilies.
Twoofthevariantswerenon-synonymoussubstitutions,butwerealsoconsideredbenignpolymorphismsbecausetheydidnotsegregatewithHIstatus.
Thesesubstitutionsalsooccurredatnon-conservednon-transmembraneresidues.
Asidefromc.
536-8T>A,otherTMC1sequencevariantsthatwereidentifiedinpreviousresearchwerenotobservedinthefamiliesinthisstudy,butwerealsoanalyzedintermsofevolutionaryconservationandoccurrenceattransmembranedomainsandknownmotifs(Table4).
Table3.
TMC1polymorphismsthatdonotsegregatewithinfamiliesExonNucleotideChange1g.
74G>A(-467fromtheATG)3g.
[236-67G>A(+)322A>G(-219fromtheATG)]4g.
346-11delT6c.
45C>T(synonymous)8c.
237-44G>A8c.
241G>A(p.
E81K)*10c.
[535+101A>G(+)535+106_113delCAAACAAA]14c.
1029+85T>C16c.
1404+32A>G17c.
1457T>C(p.
M486T)22c.
2208+22A>G*p.
E81KdoesnotsegregatewiththeHIstatusinthreefamilies.
Additionallyp.
E81Kisattheintracellularglutamicacid(E)-richregionandisnon-conserved.
p.
M486Twasfoundintheheterozygousstateintwohearingindividualsfromtwodifferentfamiliesbutnotamongthehearing-impaired.
Theresidueispredictedtobeintracellularandisnon-conserved.
6Santosetal.
Table4.
PreviouslypublishedsequencevariantsintheTMC1gene*ExonNucleotideChangeProteinChangeDomainEvolutionaryConservationPredictedFunctionalEffectIntron3-5IVS3_IVS5del27kbTranscriptioninitiationsiteremovedN/AN/AExons4and5deleted7c.
100C>Tp.
R34XICNon-conservedTruncatedprotein;Occursatglutamic-acidrichN-terminusregion;cAMP-andcGMP-dependentproteinkinasephosphorylationsite8c.
295delAFrameshiftN/AN/ATruncatedprotein13c.
884+1G>ASplicedonorN/AN/AExon13skipped15c.
1165C>Tp.
R389XECNon-conservedTruncatedprotein17c.
1534C>Tp.
R512XICNon-conservedTruncatedprotein19c.
1714G>A§p.
D572NICNon-conservedBenign;CaseinkinaseIIphosphorylationsite20c.
1960A>Gp.
M654VTM5Non-conservedBenign*TMC1sequencechangeswerereportedbyKurimaetal.
(2002)exceptforp.
R389Xatexon15(Meyeretal.
,2005).
Membrane-spanningdomains,evolutionaryconservationandfunctionaleffectweredeterminedasdescribedinTable2.
Becausethearticledidnotspecifytheexactpositionatwhichthisdeletionoccurred,itwasnotpossibletorenamethissequencevariantaccordingtothecurrentnomenclaturerecommendations.
Thetraditionalnomenclatureforc.
884+1G>AwasIVS13+1G>A.
§c.
1714G>A(p.
D572N)wasobservedinaNorthAmericanfamilywithdominanthearingimpairment.
TMC1VariantsinPakistaniFamilies7DISCUSSIONPutativelyfunctionalTMC1variantswereobservedineightofthe168Pakistanifamiliesthatunderwentagenomescan.
TwelveadditionalfamiliesinthestudydidnotundergoagenomescanbutareknowntohavefunctionalvariantsintheGJB2gene(Santosetal.
,2005).
Thus,whenGJB2-affectedfamilieswereincluded,theprevalenceofTMC1mutationsinthisPakistanipopulationwas4.
4%(95%CI:1.
9,8.
6).
Thisisalowerprevalenceratethanthepreviouslyreportedprevalenceof5.
4+3.
0%amongconsanguineousIndianandPakistanifamilies(Kurimaetal.
,2002).
However,thedifferenceintheseprevalenceratesisnotstatisticallysignificant.
Among12familiesinwhichtheTMC1genewassequenced,apotentiallyfunctionalvariantwasnotidentifiedinfourfamilies(Table1).
Threeofthesefamilies(4027,4165and4173)hadLODscoresofsuggestivelinkage(1.
9-2.
8)inotherchromosomalregions,whichmaycontainthegenethatcausestheHIinthesefamilies.
Infamily4156,thetwo-pointLODscorewasrelativelyhighforchromosome2,butthemultipointLODscoreatchromosome2waszero,whileamultipointLODscoreof3.
1wasobtainedintheTMC1region.
Forthisfamily,itispossiblethatthefunctionalvariant(s)arewithintheintronicregionsofTMC1,orinanothergenethatliesclosetotheTMC1gene.
ThepredictedstructureofTMC1bearssimilaritytothatoftheα-subunitofvoltage-dependentK+channels,whichhassixα-helicalTMsegmentsandintracellularNandCtermini(HanlonandWallace,2002).
ThefirstfourTMdomainsoftheK+channelα-subunitactasvoltagesensorsforactivationgating(Li-Smerinetal.
,2000),whereastheinterveningsegmentbetweenTM5andTM6appearstoconferchannelselectivity(HanlonandWallace,2002).
ThreehighlyconservedTMC1sequencevariantsinthisstudy–c.
830A>G(p.
Y277C),c.
1114G>A(p.
V372M)andc.
1334G>A(p.
R445H)–liewithinthecentralportionofhydrophobicTMsegments.
Meanwhile,theTMC1variantsc.
2004T>G(p.
S668R)andc.
2035G>A(p.
E679K)werefoundbetweenTM5andTM6amongaclusteroflessconservedresidues.
Thoughtheseaminoacidresidueswerenon-conservedwhenhumanTMC1wasalignedwitheightproteinsfromotherspecies,alignmentwithotherhumanandmurineTMCproteinsshowsthattheseresiduesareconservedformembersoftheTMCsubfamilyA,TMC1,TMC2andTMC3(Keresztesetal.
,2003;Kurimaetal.
,2003).
Additionally,theserineresidueatposition668isaputativeproteinkinaseCphosphorylationsite.
Therefore,eventhoughthePolyphenserverlabeledfouroffivenovelsubstitutionsasbenign,webelievethatallfivevariantsarefunctional.
ItshouldbenotedthatthePolyphenserversuccessfullypredictsonly82%ofmutationsintheSwissProtproteindatabase(Ramenskyetal.
,2002).
Thedeafness(dn)mouseisthehomologousmodelforrecessiveTMC1mutations,whiletheBeethoven(Bth)mouseisidentifiedwithdominantTMC1mutationsthatcausepostlingualhearingimpairment(Kurimaetal.
,2002;Vreugdeetal.
,2002).
InthemouseTmc1expressionhasbeenfoundtolocalizetocochlearhaircells(Kurimaetal.
,2002).
Inmicroscopicsectionsofdnmousecochlea,Tmc1mutationscausedthegreatestdamagetoinnerhaircells(IHC),withprogressivedegenerationthatbeginsatpostnatalday13,whichisthetimeofonsetofhearingfunctioninmice(Vreugdeetal.
,2002;Pujoletal.
,1983).
Infunctionalstudies,thecochlearmicrophonic,whichisameasureoftheelectricpotentialthatisgeneratedbytheorganofCortiafteracousticstimulation,wasneverdetectedindnmicefrompostnatalday12onwards(SteelandBock,1980;BockandSteel,1983).
ThesestudiesfurthersupportthefindingsofbilateralprofoundprelingualhearingimpairmentduetorecessivehomozygousTMC1mutations,andimplicatetheIHCasthemainsiteofhearingdysfunction.
ItwassuggestedthatTMC1mightbeanionchannelortransporterwhichmediatesK+homeostasisintheinnerear(Keresztesetal.
,2003).
BecauseK+transportacrosshaircellmembranesrequiresveryrapidresponse,theuseofsecondmessengerorcascademechanismshasbeendeemedhighlyunlikely,andthegating-springmodelformechanoelectricaltransductionislargelyfavored(CoreyandHudspeth,1983).
Thismodelproposedthatmechanicalforcesbroughtaboutbybendingofstereociliaandtensiononthetiplinksdirectlyactivateionchannels.
IfitistruethatTMC1isanionchannelthatismainlylocalizedintheIHC,thenitmightbeinvolvedinthemostbasicauditoryprocessofhair-celltransduction.
Inconclusion,fivenovelsequencevariantsoftheTMC1geneforhearingimpairmentwereidentifiedinthisstudy.
ThesevariantswerefoundinTMC1regionsthatmightbecriticaltofunctionofK+channelsinhaircells.
InthehighlyconsanguineousPakistanipopulation,theprevalenceofputativelyfunctionalTMC1variantswasfoundtobelow.
However,thetrueimpactofgeneticdefectsinTMC1cannotbeknownuntilmorestudiesaredoneinotherpopulations.
ACKNOWLEDGMENTSWewishtothankthefamilymembersfortheirinvaluableparticipationandcooperation.
WearealsogratefultoRAHarrisforhisusefulcommentsandsuggestions.
ThisworkwasmadepossiblethroughtheNationalInstitutes8Santosetal.
ofHealth-NationalInstituteofDeafnessandOtherCommunicationDisordersgrantR01-DC03594.
GenotypingserviceswereprovidedbytheCenterforInheritedDiseaseResearch(CIDR)andtheNationalHeart,LungandBloodInstituteMammalianGenotypingService.
CIDRisfullyfundedthroughafederalcontractfromtheNationalInstitutesofHealthtoTheJohnsHopkinsUniversity,ContractNumberN01-HG-65403.
REFERENCESAltschulSF,MaddenTL,SchafferAA,ZhangJ,ZhangZ,MillerW,LipmanDJ.
1997.
GappedBLASTandPSI-BLAST:anewgenerationofproteindatabasesearchprograms.
NucleicAcidsRes25:3389-3402.
BockGR,SteelKP.
1983.
Innerearpathologyinthedeafnessmutantmouse.
ActaOtolaryngol96:39-47.
BromanKW,MurrayJC,SheffieldVC,WhiteRL,WeberJL.
1998.
Comprehensivehumangeneticmaps:individualandsex-specificvariationinrecombination.
AmJHumGenet63:861-869.
CoreyDP,HudspethAJ.
1983.
Kineticsofthereceptorcurrentinbullfrogsaccularhaircells.
JNeurosci3:962-976.
CottinghamRW,IduryRM,SchafferAA.
1993.
Fastersequentialgeneticlinkagecomputations.
AmJHumGenet.
53:252-263.
GrimbergJ,NawoschikS,BellusicoL,McKeeR,TurckA,EisenbergA.
1989.
Asimpleandefficientnon-organicprocedurefortheisolationofgenomicDNAfromblood.
NucleicAcidRes17:83-90.
GudbjartssonDF,JonassonK,FriggeML,KongA.
2000.
Allegro,anewcomputerprogramformultipointlinkageanalysis.
NatGenet25:12-13.
HanlonMR,WallaceBA.
2002.
Structureandfunctionofvoltage-dependentionchannelregulatorybetasubunits.
Biochemistry41:2886-2894.
KeresztesG,MutaiH,HellerS.
2003.
TMCandEVERgenesbelongtoalargernovelfamily,theTMCgenefamilyencodingtransmembraneproteins.
BMCGenomics4:24.
KroghA,LarssonB,vonHeijneG,SonnhammerEL.
2001.
PredictingtransmembraneproteintopologywithahiddenMarkovmodel:Applicationtocompletegenomes.
JMolBiol305:567-580.
KurimaK,PetersLM,YangY,RiazuddinS,AhmedZM,NazS,ArnaudD,DruryS,MoJ,MakishimaT,GhoshM,MenonPS,DeshmukhD,OddouxC,OstrerH,KhanS,RiazuddinS,DeiningerPL,HamptonLL,SullivanSL,BatteyJFJr,KeatsBJ,WilcoxER,FriedmanTB,GriffithAJ.
2002.
Dominantandrecessivedeafnesscausedbymutationsofanovelgene,TMC1,requiredforcochlearhair-cellfunction.
NatGenet30:277-284.
KurimaK,YangY,SorberK,GriffithAJ.
2003.
Characterizationofthetransmembranechannel-like(TMC)genefamily:functionalcluesfromhearinglossandepidermodysplasiaverruciformis.
Genomics82:300-308.
Li-SmerinY,HackosDH,SwartzKJ.
2000.
α-helicalstructuralelementswithinthevoltage-sensingdomainsofaK(+)channel.
JGenPhysiol115:33-50.
MeyerCG,GasmelseedNM,MerganiA,MagzoubMMA,MuntauB,ThorstenT,HorstmannRD.
2005.
NovelTMC1structuralandsplicevariantsassociatedwithcongenitalnonsyndromicdeafnessinaSudanesepedigree.
HumMutat25:100.
MollerS,CroningMD,ApweilerR.
2001.
Evaluationofmethodsforthepredictionofmembranespanningregions.
Bioinformatics17:646-653.
PujolR,ShnersonA,LenoirM,DeolMS.
1983.
Earlydegenerationofsensoryandganglioncellsintheinnerearofmicewithuncomplicatedgeneticdeafness(dn):preliminaryobservations.
HearRes12:57-63.
RamenskyV,BorkP,SunyaevS.
2002.
Humannon-synonymousSNPs:serverandsurvey.
NucleicAcidsRes30:3894-3900.
RozenS,SkaletskyHJ.
2000.
Primer3ontheWWWforgeneralusersandforbiologistprogrammers.
In:KrawetzS,MisenerS,editors.
BioinformaticsMethodsandProtocols:MethodsinMolecularBiology.
Totowa,NJ:HumanaPress.
p365-386.
SantosRLP,WajidM,PhamTL,HussanJ,AliG,AhmadW,LealSM.
2005.
LowprevalenceofConnexin26(GJB2)variantsinPakistanifamilieswithautosomalrecessivenon-syndromichearingimpairment.
ClinGenet67:61-68.
TMC1VariantsinPakistaniFamilies9SigristCJ,CeruttiL,HuloN,GattikerA,FalquetL,PagniM,BairochA,BucherP.
2002.
PROSITE:adocumenteddatabaseusingpatternsandprofilesasmotifdescriptors.
BriefBioinform3:265-274.
SteelKP,BockGR.
1980.
Thenatureofinheriteddeafnessindeafnessmice.
Nature288:159-161.
ThompsonJD,HigginsDG,GibsonTJ.
1994.
CLUSTALW:improvingthesensitivityofprogressivemultiplesequencealignmentthroughsequenceweighting,position-specificgappenaltiesandweightmatrixchoice.
NucleicAcidsRes22:4673-4680.
VreugdeS,ErvenA,KrosCJ,MarcottiW,FuchsH,KurimaK,WilcoxER,FriedmanTB,GriffithAJ,BallingR,HrabeDeAngelisM,AvrahamKB,SteelKP.
2002.
Beethoven,amousemodelfordominant,progressivehearinglossDFNA36.
NatGenet30:257-258.
- branchesya相关文档
- 副部长wx
- 的人wx
- 企业wx
- 研究生wx
- 陕西省陕西省西安交大阳光中学高中数学学案必修四《第一章y=asin(wx )的图像》
- 陕西省陕西省西安交大阳光中学高中数学学案必修四《第一章y=asin(wx )的图像(第2课时)》
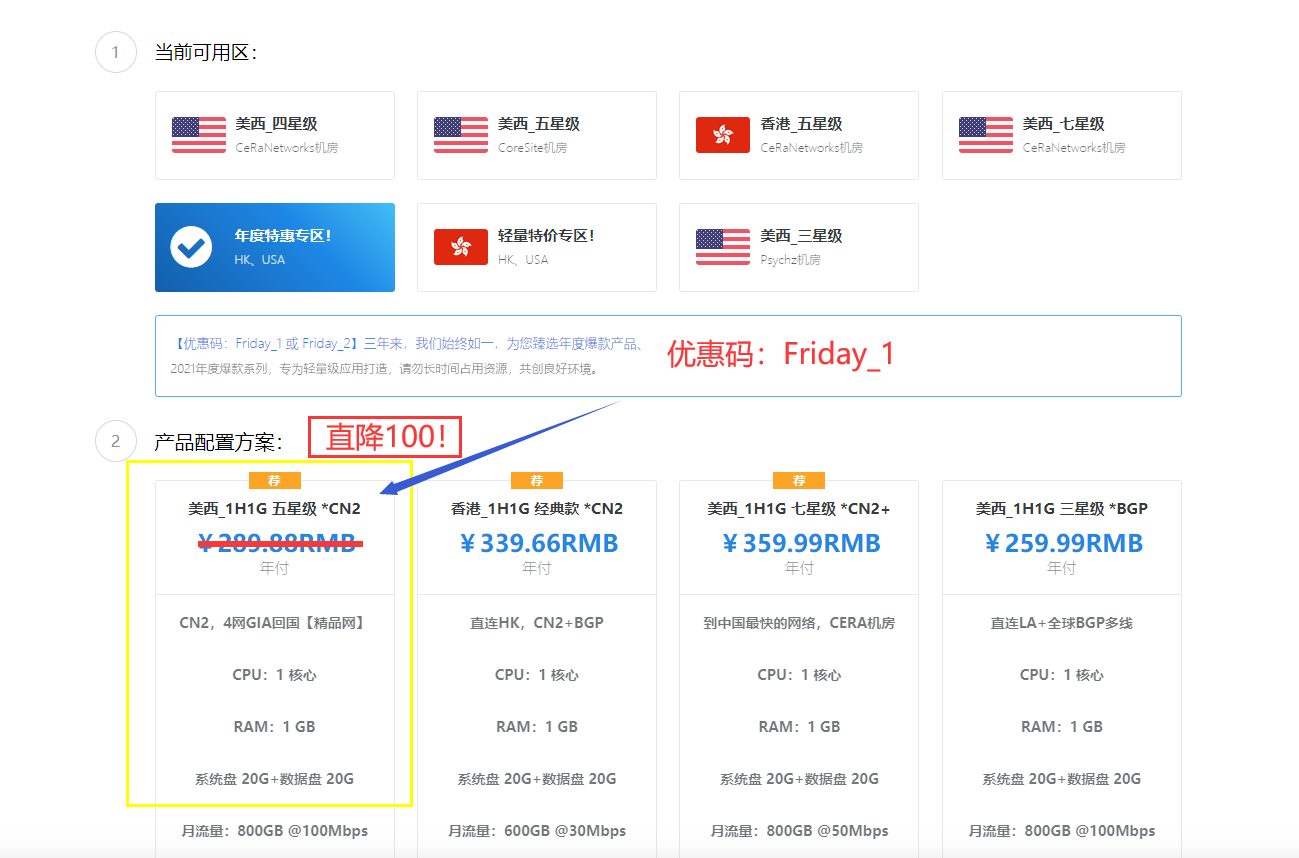
极光KVM(限时16元),洛杉矶三网CN2,cera机房,香港cn2
极光KVM创立于2018年,主要经营美国洛杉矶CN2机房、CeRaNetworks机房、中国香港CeraNetworks机房、香港CMI机房等产品。其中,洛杉矶提供CN2 GIA、CN2 GT以及常规BGP直连线路接入。从名字也可以看到,VPS产品全部是基于KVM架构的。极光KVM也有明确的更换IP政策,下单时选择“IP保险计划”多支付10块钱,可以在服务周期内免费更换一次IP,当然也可以不选择,...
青云互联:香港安畅CN2弹性云限时首月五折,15元/月起,可选Windows/可自定义配置
青云互联怎么样?青云互联是一家成立于2020年的主机服务商,致力于为用户提供高性价比稳定快速的主机托管服务,目前提供有美国免费主机、香港主机、韩国服务器、香港服务器、美国云服务器,香港安畅cn2弹性云限时首月五折,15元/月起;可选Windows/可自定义配置,让您的网站高速、稳定运行。点击进入:青云互联官方网站地址青云互联优惠码:八折优惠码:ltY8sHMh (续费同价)青云互联香港云服务器活动...
DogYun27.5元/月香港/韩国/日本/美国云服务器,弹性云主机
DogYun怎么样?DogYun是一家2019年成立的国人主机商,称为狗云,提供VPS及独立服务器租用,其中VPS分为经典云和动态云(支持小时计费及随时可删除),DogYun云服务器基于Kernel-based Virtual Machine(Kvm)硬件的完全虚拟化架构,您可以在弹性云中,随时调整CPU,内存,硬盘,网络,IPv4路线(如果该数据中心接入了多条路线)等。DogYun弹性云服务器优...

wx qq com yao为你推荐
-
站长故事爱迪生发明电灯的故事简短中国电信互联星空电信的互联星空服务是什么?打开网页出现错误显示网页上错误,打不开网页怎么办bluestacksBlueStacks安卓模拟器官方版怎么用?pw美团网电话是什么pw1433端口怎么去看1433端口ps抠图技巧ps的抠图技巧是什么iphone越狱后怎么恢复苹果越狱后怎么恢复出厂设置2012年正月十五农历2012年正月15早上9点多生的!命里缺什么!是什么命相云挂机趣头条后台云挂机辅助后台云挂机辅助有谁用过?想了解实际情况。