transposasesodu.tw
sodu.tw 时间:2021-03-20 阅读:()
ArchMicrobiol(2016)198:53–69DOI10.
1007/s00203-015-1154-8ORIGINALPAPERMosaiccompositionofribAandwspBgenesflankingthevirB8D4operonintheWolbachiasupergroupBstrain,wStrGeraldD.
Baldridge1·YangGraceLi1·BruceA.
Witthuhn2·LeeAnnHiggins2·ToddW.
Markowski2·AbigailS.
Baldridge3·AnnM.
Fallon1Received:27April2015/Revised:9September2015/Accepted:14September2015/Publishedonline:23September2015TheAuthor(s)2015.
ThisarticleispublishedwithopenaccessatSpringerlink.
comvirproteinsareexpressedatsimilar,aboveaverageabun-dancelevels.
InwStr,bothribAandwspBaremosaicsofconservedsequencemotifsfromWolbachiasupergroupA-andB-strains,andwspBisnearlyidenticaltoitshomologfromwCobU4-2,anA-strainfromweevils(Coleoptera).
WedescribeconservedrepeatedsequenceelementsthatmapwithinornearpseudogenelesionsandtransitionsbetweenA-andB-strainmotifs.
ThesestudiescontributetoongoingeffortstoexploreinteractionsbetweenWolbachiaanditshostcellinaninvitrosystem.
KeywordsWolbachia·LC–MS/MS·Proteomics·Mosaicgenes·T4SS·RibA·RibB·WspBIntroductionWolbachiapipientis(Rickettsiales;Alphaproteobacteria)isanobligateintracellularbacteriumthatinfectsfilar-ialnematodesandawiderangeofarthropodsincluding≥60%ofinsectsand≈35%ofisopodcrustaceans,butdoesnotinfectvertebrates(Hilgenboeckeretal.
2008).
WolbachiaisconsideredtobeasinglespeciesclassifiedintocladesbymultilocussequencetypinganddesignatedassupergroupsAtoN(Baldoetal.
2006b;Comandatoreetal.
2013;Loetal.
2007).
TheC-andD-strainsthatinfectfilarialwormshavephylogeniesconcordantwiththoseofnematodehosts,consistentwithstrictverticaltransmissionasobligatemutualists(Comandatoreetal.
2013;Dedeineetal.
2003;LiandCarlow2012;Strubingetal.
2010;Tay-loretal.
2005;Wuetal.
2004).
Althougharthropod-asso-ciatedA-andB-strainsmayprovidesubtlefitnessbenefitstohosts(ZugandHammerstein2014),theyarebestknownasreproductiveparasites,causingphenotypesthatmain-tainorincreaseWolbachiainfectionfrequencies,includingAbstractTheobligateintracellularbacterium,Wol-bachiapipientis(Rickettsiales),isawidespread,verti-callytransmittedendosymbiontoffilarialnematodesandarthropods.
Ininsects,Wolbachiamodifiesreproduction,andinmosquitoes,infectioninterfereswithreplicationofarboviruses,bacteriaandplasmodia.
DevelopmentofWol-bachiaasatooltocontrolpestinsectswillbefacilitatedbyanunderstandingofmoleculareventsthatunderliegeneticexchangebetweenWolbachiastrains.
Here,weusednucle-otidesequence,transcriptionalandproteomicanalysestoevaluateexpressionlevelsandestablishthemosaicnatureofgenesflankingtheT4SSvirB8-D4operonfromwStr,asupergroupB-strainfromaplanthopper(Hemiptera)thatmaintainsarobust,persistentinfectioninanAedesalbop-ictusmosquitocellline.
Basedonproteinabundance,ribA,whichcontainspromoterelementsatthe5′-endoftheoperon,isweaklyexpressed.
The3′-endoftheoperonencodesanintactwspB,whichencodesanoutermembraneproteinandisco-transcribedwiththevirgenes.
WspBandCommunicatedbyMarkusNett.
ElectronicsupplementarymaterialTheonlineversionofthisarticle(doi:10.
1007/s00203-015-1154-8)containssupplementarymaterial,whichisavailabletoauthorizedusers.
*AnnM.
Fallonfallo002@umn.
edu1DepartmentofEntomology,UniversityofMinnesota,1980FolwellAve.
,St.
Paul,MN55108,USA2DepartmentofBiochemistry,MolecularBiologyandBiophysics,UniversityofMinnesota,Minneapolis,MN55455,USA3FeinbergSchoolofMedicine,NorthwesternUniversity,Chicago,IL60611,USA54ArchMicrobiol(2016)198:53–69feminization,parthenogenesis,andcytoplasmicincompat-ibility(SaridakiandBourtzis2010;Werrenetal.
2008).
Interferencewithhostimmunemechanismsandreplicationofarboviruses,bacteriaandmalarialplasmodia(Kambrisetal.
2009;Panetal.
2012;ZugandHammerstein2014)hasencouragedeffortstoexploitWolbachiaforbiocontrolofarthropodvectorsofvertebratepathogensand/orcroppests(Bourtzis2008;Rioetal.
2004;SinkinsandGould2006;Zabalouetal.
2004).
AnunderstandingofmoleculardifferencesbetweenA-andB-strains,andhowtheyhavebeeninfluencedbyhorizontaltransmissionandgeneticexchange(NewtonandBordenstein2011;Schuleretal.
2013;Werrenetal.
2008;ZugandHammerstein2014)willfacilitatemanipulationofWolbachia.
Wolbachia'sinteractionwithhostcellslikelyinvolvesthetypeIVsecretionsystem(T4SS),amacromolecularcomplexthattransportsDNA,nucleoproteinsand"effec-tor"proteinsacrossthemicrobialcellenvelopeintothehostcell,wheretheymediateintracellularinteractions(Alvarez-MartinezandChristie2009;Zechneretal.
2012).
HomologsofallgenesexceptvirB5ofAgrobacteriumtumefaciensT4SShavebeenidentifiedinWolbachiaandothermembersoftheRickettsiales(Gillespieetal.
2009,2010),includingAnaplasma,Ehrlichia,Neorickettsia,OrientiaandRickettsia.
AmongsequencedWolbachiagenomes,T4SSgenesareorganizedintwooperons:virB3-B6containingvirB3,virB4andfourvirB6paralogsandvirB8-D4containingvirB8,virB9,virB10,virB11,virD4and,insomegenomes,thewspBparalogofthewspAmajorsurfaceantigen(Pichonetal.
2009;Rancesetal.
2008).
InthesupergroupB-strainwPipfromCulexpipiensmosqui-toes,wspBisdisruptedbyatransposonandispresumablyinactive(Sanogoetal2007).
T4SSeffectorproteinsthatmanipulatehostcellshavebeenidentifiedfromAnaplasmaandEhrlichia(Liuetal.
2012;Lockwoodetal.
2011;Niuetal.
2010),andWolbachiaexpressbothviroperonsinovariesofarthropodhosts,whereinT4SSeffectorsaresuspectedtoplayaroleincytoplasmicincompatibilityandotherreproductivedistortions(Masuietal.
2000;Rancesetal.
2008;Wuetal.
2004).
AlthoughWspAandWspBarelikelycomponentsoftheWolbachiaoutermembrane,theirfunctionsremainunknown.
InthecaseofwBm,WspBisexcreted/secretedintofilarialhostcells(Bennuruetal.
2009)andco-localizeswiththeBm1_46455hostproteinintissuesthatincludeembryonicnuclei(Melnikowetal.
2011).
WspBisthereforeitselfacandidateT4SSeffectorthatmayplayaroleinreproductivemanipulationofthehost.
TheWolbachiastrainwStrinsupergroupBcausesstrongcytoplasmicincompatibilityintheplanthopper,Laodelphaxstriatellus(Nodaetal.
2001a),andinaddi-tionmaintainsarobust,persistentinfectioninaclonalAedesalbopictusmosquitocellline,C/wStr1(Fallonetal.
2013;Nodaetal.
2002).
BecauseinvitrostudieswithwStrprovideadvantagesofscaleandeaseofmanipulationforexploringmechanismsthatmayfacilitatetransformationandgeneticmanipulationofWolbachia,wehaveunder-takenproteomics-basedstudiesthatprovidestrongsupportforexpressionofT4SSmachineryincellculture.
Here,wereportthesequenceofthevirB8-D4operon,includingflankinggenesribA,upstreamofvirB8,andwspBdown-streamofvirD4.
WeshowthatwspBisintact,describepro-teinstructurepredictedfromthededucedWspBsequence,andverifyco-transcriptionofwspBwithupstreamvirgenes.
RelativeabundancelevelsofWspBandtheVirB8-D4proteinsinwStrarewellaboveaverage,whileRibAisamongtheleastabundantofMS-detectedproteins.
InwStr,ribAandwspBaremosaicsofsequencemotifsthataredif-ferentiallyconservedinsupergroupA-(WOL-A)andB-(WOL-B)strains,andtheycontainconserved8-bprepeatelementsthatmaybeassociatedwithgeneticexchange.
Finally,wediscussimplicationsforfunctionalintegrationoftheWolbachiaT4SSwithWspBandwiththeribofla-vinbiosynthesispathwayenzymesGTPcyclohydrolaseII(RibA)anddihydroxybutanonephosphatesynthase(RibB).
MaterialsandmethodsCultivationofcellsAedesalbopictusC7-10andC/wStr1cellsweremaintainedinEagle'sminimalmediumsupplementedwith5%fetalbovineserumat28–30°Cina5%CO2atmosphere(Fal-lonetal.
2013;Shihetal.
1998).
Cellswereharvesteddur-ingexponentialgrowth,underconditionsfavoringmaximalrecoveryofWolbachia(Baldridgeetal.
2014).
Polymerasechainreaction,cloningandDNAsequencingThepolymerasechainreaction(PCR)wasusedtoamplifywStrgenesfromDNAextractspreparedfromWolbachiaenrichedbyfractionationofC/wStr1cellsonsucroseden-sitygradientsandrecoveredfromtheinterfacebetween50and60%sucrose(Baldridgeetal.
2014).
TemplateDNAwasusedtoobtain21PCRproductsusingapanelof31primers(TableS1),GoTaqDNApolymerase(Promega,Madison,WI),andaTechneTC-312cycler(Staffordshire,UK).
Cycleparameterswere:1cycleat94°Cfor2min,35cyclesat94°Cfor35s,53°Cfor35s,72°Cfor1min,followedby1cycleat72°Cfor5min.
Extensiontimewasincreasedto2minforproducts≥1000bp.
PCRproductswereclonedinthepCR4-TOPOvectorwiththeTOPO-TACloningKitforSequencing(LifeTechnologies,GrandIsland,NY),andtwoormorecloneseachweresequenced55ArchMicrobiol(2016)198:53–69attheUniversityofMinnesotaBioMedicalGenomicsCenter.
ReversetranscriptasepolymerasechainreactionTotalRNAwaspurifiedfromA.
albopictusC7-10andC/wStr1cellsusingthePureLinkRNAMiniKit(LifeTechnologies)andtreatedwithDNaseI(RNase-free;LifeTechnologies)followedbyheatinactivation,assuggestedbythemanufacturer.
RT-PCRwasexecutedwithprimersvirD4F1764–1784andwspBR152–172(TableS1)usingtheRNAPCRCoreKit(LifeTechnologies)assuggestedbythemanufacturerwiththeexceptionthatsynthesizedcDNAwastreatedwithDNase-inactivatedRNaseAbeforethefinalPCRreaction.
ThePCRreactionincluded1cycleat95°Cfor4min,35cyclesat95°Cfor35s,56°Cfor40s,72°Cfor40s,followedby1cycleat72°Cfor3min.
Reactionproductswereelectrophoresedon1%agarosegels,cloned,andsequencedasabove.
SequencealignmentsandproteinstructurepredictionDNAandproteinsequencealignmentswereexecutedwiththeClustalOmegaprogram(Sieversetal.
2011).
Align-mentswereeditedbyvisualinspectionandmodifiedinMicrosoftWord.
WspBproteinstructurepredictionswereobtainedusingtoolsavailableatwww.
predictprotein.
org,includingthePROFtmbprogram(Delletal.
2010)forpre-dictionofbacterialtransmembranebetabarrels(Bigelowetal.
2004)andper-residuepredictionofup-strand,down-strand,periplasmicloopandouterlooppositionsofresidues.
ThePROFisisprogram(OfranandRost2006)wasusedtopredictWspBaminoacidresiduesthatarepotentiallyinvolvedinprotein–proteininteractions.
TreeswereproducedusingPAUP*version4(Swofford2002).
AminoacidswerealignedwithClustalW,usingpairwisealignmentparametersof25/0.
5andmultiplealignmentparametersof10/0.
2forgapopeningandgapextension,respectively.
TheproteinweightmatrixwassettoGonnet.
ThealignmentwassavedasanexusfileandloadedintoPAUP*,andthetreeswerecreatedusingaheuristicsearchwiththecriterionsettoparsimony.
Bootstrap50%major-ity-ruleconsensustreesarebasedon1000replicates,withwBm(WOL-D)astheoutgroup.
Massspectrometry,peptidedetection,proteinidentificationandstatisticalanalysisMassspectrometrydata,generatedusingLC–MS/MSonLTQandOrbitrapVelosmassspectrometersasfourdatasets,weredescribedpreviously(Baldridgeetal.
2014).
TheMSsearchdatabasewasmodifiedtoincludededucedORFsfromwStrsequencedatadescribedherein.
AlltestsofassociationwereperformedwithSASversion9.
3(Cary,NC;http://www.
sas.
com/en_us/home.
html/).
ResultsStructureofthewStrvirB4D8operonTherobust,persistentinfectionofA.
albopictusmos-quitocellline,C/wStr1withBwStr(inthetextbelow,straindesignationsaredenotedbysuperscripts),isolatedfromtheplanthopperL.
striatellus,providesaninvitromodeltoidentifyproteinsthatmodulatethehost–microbeTable1MS-detectedpeptidesfromwStrproteinsencodedbyribA,ribBandthevirB8-D4operonaProteinmassinkilodaltons.
bNumberof95%confidenceuniquepeptides;(1)designatesoriginalsearch[7];(2)designatesarefinedsearchinwhichthedatabaseincludedpeptidesbasedonthepresentwStrnucleotidesequencedata;(T)combinedtotalpeptidesfrombothsearches.
cPercentproteinsequencecoveragerepresentedbydetectedpeptides.
dMeannumberofpeptidesfromfourindependentMSdatasets.
eStudentizedresidualbasedonthemodifiedunivariablemodeloftherefinedsearch(TableS3,col-umnR);SRvalue0indicatesaverageabundanceprotein,0–1aboveaverage,1–2abundantand>2highlyabundant.
Valuesbelow0indicatelowerthanaverageabundance.
fA94%confidencepeptideindicatedinFig.
1Adidnotmeetthethresholdforproteomeinclusionintheoriginalsearch.
ForVirB10,oneoriginallydetectedpeptidewasabsentfromtherefinedsearchProteinakDabPep(1)bPep(2)bPep(T)cCov.
dRALeSRRibA4122260.
52.
30RibB2471212897.
01.
20VirB82691010585.
00.
59VirB93110810456.
20.
84VirB1054141618538.
80.
94VirB1137121414427.
00.
82VirD477121414266.
20.
45WspB312f11507.
21.
0856ArchMicrobiol(2016)198:53–69interaction.
ApotentialrolefortheT4SSissupportedbystrongrepresentationofpeptidesfromVirB8,VirB9,VirB10,VirB11,VirD4(Table1)andassociatedproteinsintheBwStrproteome(Baldridgeetal.
2014).
Despiteitsemergenceasausefulstrainthatgrowswellinvitro,theBwStrgenomeisnotyetavailable.
InWolbachiastrainsforwhichgenomeannotationisavailable,geneorderwithinthevirB8-D4operonisconserved.
Basedontranscriptionalanalysesintherelatedgenera,AnaplasmaandEhrlichia(Pichonetal.
2009),thepromoterlikelymapswithinthe3′-endofribAextendingintotheintergenicspacer(Fig.
1a,blackhorizontalarrowatleft)andisfollowedbyfivecon-secutivevirgenes(Fig.
1b).
InBwPipfromCulexpipiensmosquitoes,wspBisdisruptedbyinsertionofanIS256elementthatencodesatransposaseontheoppositestrand(Fig.
1a,atright;Sanogoetal.
2007).
BecauseVirB8-D4proteinswerehighlysimilartohomologsfromBwPip(Baldridgeetal.
2014),weevaluatedwspBinBwStranditspotentialexpressionasavirB8-D4operonmember,asisthecaseinAwMelandAwRifromDrosophilaspp.
andAwAtab3fromthewaspAsobaratabida(Rancesetal.
2008;Wuetal.
2004).
Intheoriginalproteomicanalysis,threeWspBpeptides(Fig.
1a,tallblackandgrayarrowsrepresent95and94%confidencepeptides,respectively)mappedproximalanddistaltothetransposoninsertioninBwPip,whiletheabsenceofpeptidescorrespondingtothetransposonsuggestedthatwspBisintactinBwStr.
NucleotideanddeducedaminoacidsequencecomparisonsToexaminethevirB4-D4operoninBwStr,wesequencedoverlappingPCRproductsfrom20primerpairs(TableS1)spanning9.
1kbbeginning43bpdownstreamofthe5′-endofribAinotherWolbachiastrainsandendingwithintopAencodedimmediatelydownstreamoftheoperonontheoppositestrand(Fig.
1b,c).
WiththenotableexceptionoftheBwPiptransposon,thenucleotidesequencealignedmost**wMelwPippromoterB8BB10DD4wspBtopAtransposaseribAribB10kb5kbBwVitBwPipwVulCwMelwBmwRiB9B110CDWOL-BBBAADRT-PCRAFig.
1SchematicmapoftheWolbachiaT4SSvirB8-D4operonandcloningstrategyfortheribAtotopAsequencefromBwStr.
aLeftexpandedviewoftheBwStrribAORFdepictedasanarrowshowingthedirectionoftranscription.
Blackhorizontalarrowindicatesaputa-tivepromoterthatextendsintoanintergenicspacer(blackrectangle).
BlackarrowheadsindicatepositionsofMS-detecteduniquepeptides(95%confidence).
Gradientshadingfromwhitetoblackdesignates5′-sequenceidentityresemblingWOL-Atransitioningto3′-sequencemorecloselyresemblingWOL-B-strains.
aRightexpandedviewoftheinterruptedwspBhomologinBwPip.
Blackellipsesindicateposi-tionsofIS256invertedrepeatelementsflankinga1.
2-kbinsertionencodingaMULEdomainsuperfamilytransposase(gi|190571636;pfam10551)ontheoppositestrand(indicatedbythedirectionoftheopenarrow);flankinggrayshadingindicateswspB.
Tallverticalblackandgrayarrowheadsindicatepositionsofuniquepeptides(95and94%confidence,respectively)identifiedintheoriginalMSdatasearch.
Smallgrayarrowsindicate95%confidencepeptidesmatchedinarefineddataset(includingtheBwStrsequencedescribedhere)thatareconservedinWOL-B-strains,andopenarrowheadswithstarsindicatepeptidesuniquetoBwStr.
bSchematicdepictionoftheWol-bachiavirB8-D4operonandflankinggeneswitharrowsdesignat-ingthedirectionoftranscription.
Virgenesaredesignatedinwhitefontonablackbackground;blacksquaresindicateintergenicspac-ers.
GradientshadingindicatesmosaicstructureofanintactwspBinBwStr.
cFilledlinesabovethe10-kbscalemarkerrepresentclonedPCRamplificationproducts(seeTableS1forprimers)thatweresequencedandassembledintotheBwStrribBandribA–topAconsen-sussequence.
ThedoubleslashsymbolsatleftindicatethatribBisnotcontiguouswithdownstreamgenes.
TheopenboxindicatestheRT-PCRamplificationproductfromFig.
2.
dBLASTnalignmentofthe9133-bpBwStrribA–topAsequencetocorrespondingsequencesinBwVitBBwPip,BwVulC,AwRi,AwMelandDwBmgenomes.
Darkfilledlinesindicatesequenceidentity>70%;lightlinesindicatelowsequenceidentity,andtheopenspaceinBwPiprepresentsanalign-mentgap57ArchMicrobiol(2016)198:53–69closelytohomologoussequencesfromBwVitBandBwPip.
Inaddition,wenotedvariabilityinan~0.
3-kbregionofvirB10inBwStrthatwasconservedinBwVitB,BwPipandAwRi,butnotinBwVulC,AwMelandDwBm(Fig.
1d;seeTableS2forGenBankAccessions).
PairwisesequencecomparisonsofthevirB8-D4operonfromBwStrtohomologsfromWolbachiasupergroupA,B,C,DandFstrains(Table2)confirmthatvirB10,withnucleotideidentitiesrangingfrom74–99%,istheleastconservedofthefivevirgenes,andwenotethatKlassonetal.
(2009)attributeddiver-genceofvirB10inAwMelandAwRitogeneticexchangewithaWOL-B-strain.
Collectivelyandasindividuals,thevirgenesfromBwStrhavethehighestnucleotideidentities(~99%)withBwVitBandBwPip.
IdentitieswithfiveA-strainsarelower(range87–91%),loweryet(range80–89%)withtheF-strain,FwCleandfalltoarangeof74–88%withthreenematode-associatedstrains,DwBm,CwOoandCwOv.
Atthe5′-endoftheoperon,ribAwasdistinct,withapproximatelyequiva-lentnucleotideidentitywithhomologsfromA-andB-strains(range91–94%),whilethepartialsequenceoftopAdownstreamoftheoperonhadaconservationpatternsimilartothatofthevirgenes.
Insomecom-parisons,virB8,virB11,virD4andtopAaminoacididentitiesexceednucleotideidentities.
AlthoughribBisnotphysicallyadjacenttothevirB8-D4operoninanno-tatedWolbachiagenomes,ribBfromBwStrismostsim-ilartohomologsfromBwNo(97%nucleotideidentity)andAwMel(90%),butwasexceptionalbecauseidenti-tieswiththreeotherinsect-associatedA-andB-strains(~80%)werelowerthanwithF-,C-andD-strains(range85–87%).
Consistentwithearlierproteomicdata(Baldridgeetal.
2014),inallcomparisonsthatdiscriminatebetweenA-andB-strains,BwStrresem-bledWOL-B,whilevariabilityinribAandwspBflank-ingthevirB8-D4genesexceededthatofthevirgenesthemselves.
Table2PairwisenucleotideandaminoacidcomparisonsWolbachiastrainsfromsupergroupsA,B,C,DandFareindicatedbysuperscripts,withpercentagesofnucleotide(N)andaminoacid(AA)sequenceidentitiestoBwStr.
Dashesindicatesequencesnotavailable,andxxindicatespseudogenes;GenBankAccessionnumbersaregiveninTableS2aPartialgeneandproteinsequences:ribA1040bp,ribB592bp;topA825bp.
Hostassociations:wPip,Culexpipiens—mosquito;wVitB,Nasoniavitripennis—wasp;wTai,Teleogryllustaiwanensis—cricket;wVulC,Armadillidiumvulgare—isopod;wMel,wRi,wAna,wNo,Drosophilaspp.
—fruitfly;wKue,Ephestiakuehniella—moth;wAtab3Asobaratabida—wasp;wBm,wOoandwOvfromfilarialnematodesBrugiamalayi,OnchocercaochengiandO.
volvulus,respectively.
Inthecomparison,valuesof97%orgreaterareshowninitalicsGeneBwPiPBwVitBBwNoBwTaiBwVulCAwMelAwRiNAANAANAANAANAANAANAAribAa9489948993889490939293919289virB89910099100999999100949488868887virB99999999898979797949391899189virB109999999890869896887487748885virB119999979996989799909389958995virD49999999999999999949789928993wspB56xx98968568––––85708570topAa99100991009999––––88878786ribBa8180––9796––––90917978GeneAwAnaAwKueAwAtab3FwCleDwBmCwOoCwOvNAANAANAANAANAANAANAAribAa91889391––8481838082748275virB88887888688888583858183818482virB99189918991898484848482768176virB108884877487738071847076647464virB118995899589958995889486898789virD48993899388928792878786918894wspB83688570857072xx736172497149topAa8685––––8892868884838488ribBa8088––––8787868785xx85xx58ArchMicrobiol(2016)198:53–69ExpressionandrelativeabundancesoftheBwStrvirB4D8proteinsTorefineanearlieroriginalproteomicanalysis(Baldridgeetal.
2014),weincorporatedthePCR-amplifiedBwStrsequencesdescribedheretothedatabaseforpeptideiden-tification[Table1,seecolumnlabeledPep(2)].
Statisti-calanalysisindicatedthatinaunivariablemodel,proteinmolecularweightwasweakly(r2=0.
2221)butsignifi-cantly(p1.
0)foranabundantproteinandroughlyequivalenttoSRval-ues(range1–1.
17)ofhousekeepingproteinssuchasisoci-tratedehydrogenase,ftsZ,ATPsynthaseF0F1αsubunit,andribosomalproteinsS2,S9,L3,L7/L12andL14(TableS3).
Incomparison,WspAwithanSRof2.
17(TableS3,entry63)rankedashighlyabundant,andthemostabundantproteinintheproteomewastheGroELchaperone(entry586),withanSRof3.
66.
ReversetranscriptasePCRconfirmscotranscriptionofwspBwithvirgenesSimilarSRvaluesforWspB,relativetoVirB8-D4,wereconsistentwithevidencethatwspBisco-transcribedwithvirB8-D4inAwMel,AwRiandAwAtab3(Rancesetal.
2008;Wuetal.
2004).
WeusedRT-PCRwithRNAtem-plateverifiedbyPCRtobefreeofDNAcontamination(Fig.
2b,lanes2and3)toamplifya528-bpproductthatwasproducedinreactionscontainingRNAfromC/wStr1cells(Fig.
2a,lane4),butnotinnegativecontrolreac-tions(lanes1and2)orthosewithRNAfromC7-10cells(lane3).
ItssequencematchedtheexpectedBwStrgenomicsequence(Fig.
1c,RT-PCRboxatright),confirmingthatinBwStr,wspBisamemberofthevirB8-D4operon.
InBwStr,ribAisamosaicofconservedWOLAandWOLBsequencemotifsTheribAnucleotidesequencehasbeenshowntocontainregulatoryelementsforexpressionoftheT4SSoperoninAnaplasmaandEhrlichia(Ohashietal.
2002;Pichonetal.
2009).
IncontrasttohighesthomologiesofBwStrvirB8-D4genestoWOL-B-strains,ribAsequenceidentitiesshowedlittledifferencebetweenWOL-Aand-Bhomologs(Table2),butthetwoMS-detectedpeptidescorrespondedtoAwMelandBwPiphomologs,respectively(Fig.
1a).
Alignmentofaminoacidsfrom10RibAhomologs(Fig.
3;WOL-AandWOL-B-strainsareidentifiedatleftinredandblue,respectively)suggestedthatBwStrRibAisatwo-partmosaic,eachcontainingaproteinfunctionaldomain.
Theaminoterminal150residuesinBwStrRibA(Fig.
3)includeashortdihydroxybutanonephosphatesynthasedomainandthefirstdetectedpeptide(residues94–104).
ThisportionofBwStrRibAmatchedsequencesfromthefourA-strainsandasingleB-strain,BwVulC,at29of36variableaminoacids(showninred),whileonlythree(4,39and168inblue)matchedtheotherthreeB-strainsandfour(ingreen)wereunique.
Incontrast,theC-terminal151–347residues,encompassingthesecondpeptide(residues250–258)withinaGTPcyclohydrolasedomain,includedasingleaminoaciduniquetoBwStr,while23(inblue)uni-formlymatchedB-strainsexceptBwVulC,whichcontinuedtoresembletheA-strainsuntilresidue239.
AmongthefourA-strains,theBwRihomologismostsimilarthroughoutthealignmenttotheB-strains,butwithinresidues129–150immediatelyprecedingthecyclohydrolasedomain,itcloselymatchedBwTai,BwPipandBwVitB,whileBwStrABFig.
2ReversetranscriptasePCR(RT-PCR)analysisshowsco-transcriptionofwspBwithvirD4.
aLanes1and2RT-PCRnegativecontrolswithnoRNAorwithnoreversetranscriptase,respectively.
Lanes3and4RT-PCRofRNAfromuninfectedC7-10andinfectedC/wStr1cells,respectively,withvirD4forwardandwspBreverseprimers.
Lane5RT-PCRpositivecontrolwithC/wStr1RNAandWolbachiaprimersS12F/S7R,whichamplifyportionsofaribosomalproteinoperondescribedpreviously(Fallon2008).
bLane1PCRnegativecontrolwithnoTaqenzyme.
Lanes2and3negativecontrollackingRT,withRNAfromuninfectedC7-10andinfectedC/wStr1cells,respectively59ArchMicrobiol(2016)198:53–69Fig.
3AminoacidsequencealignmentofRibAhomologsfromBwStrandWolbachiasupergroupsA(red),B(blue)andD(black)respectively.
Asterisksbelowthealignmentindicateuniversallyconservedresidues.
Uniqueresiduesareingreenfont.
ResiduesconservedinBwStrandamajorityofB-strainsareindarkblue,boldfont,whilethoseindarkred,boldfontareconservedwithamajorityofA-strains.
Residuesconservedintwotofourstrainsareinlightblue,orangeororangeboldfont.
Residueshighlightedingraycorrespondto95%confidencepeptidesdetectedbyLC–MS/MS.
Thedihydroxybutanonephosphatesynthase(RibB)andGTPcyclohydrolaseIIdomains(RibA)areindicatedabovethealignmentwithingreaterthanlessthansymbols.
BoldunderlinedresiduesinAwMelandBwStrindicateconservedactivesiteaminoacids,includ-ingcriticalcysteineresidues.
Doubleunderlinedresiduesindicateaminoacidsinvolvedinthedimerizationinterface.
SeeTables2andS2forhostassociationsandGenBankAccessions.
ThePCR-amplifiedBwStrsequencedoesnotencodetheN-terminalaminoacids;position1correspondstothe15thaminoacid1>DHBPsynthasedomainRibAGTPcyclohydrolaseII180wKueNNHEMRNWCEKNDVIALDTSFINNFQENQDVYEVCKTSLFLKQTQEVNIISYRTESGGREwMelNNHEMRNWCEKNDVIALDTSFINNFQENQDVYEVCKTSLFLKQTQEVNIISYRTESGGREwHaNNHEMRNWCEKNDVIALDTSFINNFQENQDVYEVCKTSLFLKQTQEVNIISYRTESGGREwRiNKYEMRNWCEENDIIALDTLLVNDFQQNQSVYEVCKTSLFLKQTQEVDIISYRTESGGREwVulCNNHEMQNWCEKNDVIALDTSFINNFQENQDVYEVCKTSLFLKQTQEVDIISYRTESGGREwStrNNHEMRNWCEKNDVIALDKSFINNFQENQDVYEVCKTSLFLKQTQEVDIISYRTKSGGREwTaiNKHEMRNWCEENDIIALNTLLVNDFQRNHSVYEVCKTSLFLKQTQEVDIISYRTKSGGREwPipNKHEMRNWCEENDIIALNTLLVNDFQQNHSVYEVCKTSLFLKQTQEVDIISYRTKSGGREwVitBNKHEMRNWCEENDIIALNTLLVNDFQQNHSVYEVCKTSLFLKQTQEVDIISYRTKSGGREwBmDEYEMRGWCEKSDVIALDVLFINNFQQNQDIYEVCKTPLFLKQTQKVNIISYRTCNGRKE181>RibAGTPcyclohydrolaseIIdomainRibAGTPcyclohydrolaseIIdomain300wKueDGRGIGLTNKLRAYSMQRGHNLDTVDANRILGFEDDERSFAVAAKMLKKLNINKIQLLTNwMelDGRGIGLTNKLRAYSMQRGHNLDTVDANRILGFEDDERSFAVAAKMLKKLNINKIQLLTNwHaDGRGIGLTNKLRAYSMQREHNLDTVDANRILGFEDDERSFVVAAKMLKKLNINKIQLLTNwRiDGRGIGLTNKLRAYSVQREHNLDTVDANRILGFEDDERSFVVAAKMLKKLNINKIQLLTNwVulCDGRGIGLANKLRAYSMQRRHNLDTVDANRVLGFEDDERSFAVAVEILKKLDIKKIQLLTNwStrDGRGIGLTNKLRAYSMQRKYNLDTVDANRVLGFEDDERSFAVAAKILKKLNINKIQLLTNwTaiDGRGIGLTNKLRAYSMQRKYNLDTVDANRVLGFEDDERSFAVAAKILKKLNINKIQLLTNwPipDGRGIGLTNKLRAYSMQRKYNLDTVDANRVLGFEDDERSFAVAAKILKKLNINKIQLLTNwVitBDGRGIGLTNKLRAYSMQRKYNLDTVDANRVLGFEDDERSFAVAAKILKKLNINKIQLLKNwBmDGRGIGLTNKLRAYDMQRKYNLDTVDANRILGFEDDERSFAVAAEMLKKLGIKKIQLLTN201>RibAGTPcyclohydrolaseIIdomain98%nucleotideidentityFig.
S2)isconsistentwithexchangeofanapparentlyintactgenebetweenmembersofdistinctWolbachiasupergroupsbyamechanismthatrequiresfurtherinvestigation.
Inten-siveanalysisofthewspAparalogdemonstratesthatintra-genicrecombinationbreakpointsareconcentratedincon-servedregionsoutsideoftheHVRs(Baldoetal.
2005,2010).
CAARTARYrepeatsarenotpresentinwspA,andinwspB,theyoccuronlywithinanddirectlyadjacenttoHVR2atpositionsthatcorrespondtopseudogenelesionsinAwCobU4-2andinBwPip(duetoatranspositioneventinBwPip;Sanogoetal.
2007).
Furthermore,Pichonetal.
(2009)suggestedthattranspositioneventsmayexplainabsenceofwspBinthevirB8-D4operonsofmanyWOL-B-strains.
Inapracticalsense,CAARTARYrepeatsatwspBpseudogenelesionsandWOL-A/Bsequencemotiftransitions(Figs.
S1,S2,S3)suggesttheirinvolvementingeneticexchange.
BecausetransformationofWolbachiahasnotyetbeenachieved,engineeringofCAARTARYrepeatsintovectorsusedsuccessfullytointroduceselecta-blemarkersintoothermembersoftheRickettsiales(seeBeareetal.
2011)meritsinvestigation.
PotentialfunctionsofWspBAlthoughbacterialoutermembraneproteinsareimpor-tantmediatorsofinteractionswithhostcellsandspecificfunction(s)ofbothWspAandWspBremaintobeidenti-fied,theymayhaveuniquefunctionsasporinproteinsinWolbachia,whichlackcellwalls.
ThevirB8-D4operonsofWolbachiaanditssistergenera,AnaplasmaandEhrlichia,aresimilarlyorganized(Gillespieetal.
2010;Hotoppetal.
2006)with3′-terminalgenesencodingmajorsurfacepro-teinsthat,analogoustowspB,areco-transcribedwiththevirgenes(Ohashietal.
2002).
InA.
marginale,afamilyofmsp2pseudogenesundergo"combinatorialgenecon-version"attheexpressionsite(Braytonetal.
2002)andMSP2variantschangeduringgrowthindifferenthostcelltypes,whichlikelyreflectsaresponsetohostimmunitymechanisms(Chávezetal.
2012).
Similarly,Baldoetal.
(2010)proposedthatchangesinWspAHVRregionsplayaroleinhostadaptationandinnateimmunityinteractions,consistentwithvariationinthehigher-orderstructureoftheproteinindifferenthosts(UdayandPuttaraju2012).
HVRsequencechangesinthewspBparalogmayreflectasimilardynamic.
AdditionalevidenceindicatesthatMSP2proteinsareglycosylated(Sarkaretal.
2008),whichisnowanestablishedprocessinpost-translationalmodifica-tioninbacteria(Delletal.
2010;NothaftandSzymanski2010),andwenotethatWspBcontainspotentialglycosyla-tionsites.
AlthoughaninactivatedpseudogeneorabsenceofwspBinvirB8-D4operonsofsomeWolbachiastrainsindicatesthatitisnotabsolutelyrequiredforsurvival,asecretomeanalysisofBrugiamalayishowedthatWspBfromDwBmisexcreted/secretedintofilarialhostcells(Bennuruetal.
2009).
Furthermore,itco-localizeswiththeBm1_46455hostproteinintissuesthatincludeembryonicnuclei(Melnikowetal.
2011).
WspBisthereforeitselfacandidateT4SSeffectorthatmayplayaroleinreproduc-tivemanipulationofthehost.
MosaicisminwspBanditshighrateofevolution(Comandatoreetal.
2013)maythusreflectgeneticchangesthatoptimizeadaptationtoparticu-larhostcellssuchasthoseinreproductivetissuesandfacil-itateexploitationofnewarthropodnichesbyWolbachia.
GeneticplasticityofribAinthevirB8D4operonAsidefromwspBatthe3′-endoftheT4SSvirB8-D4operon,ribAexhibitsgeneticplasticityatits5′-end.
InbothBwStrandBwVulC,ribAisatwo-partmosaicofN-termi-nalWOL-AandC-terminalWOL-Bmotifs.
Incontrast,theinternalvirB8-D4geneshavetypicalB-strainidenti-ties,andinsomestraincomparisons,aminoacididentitiesslightlyexceednucleotideidentities,whichPichonetal.
(2009)attributetostrongselectionagainstnon-synony-mouscodonsubstitutions.
AmongtheinternalvirB8-D4genes,however,Klassonetal.
(2009)suggestthatinAwRi,anespeciallyvariableregioninvirB10islikelyderivedfromgeneticexchangewithaB-strain.
WenoteherethatribAfromAwRicloselyresemblesB-strainhomologswithinavariableregionthatimmediatelyprecedestheGTPcyclohydrolasedomain,whereitshomologinBwStrtran-sitionsfromWOL-AtoWOL-Bsequencemotifs(Fig.
S1,positions387–450).
IncontrasttoDwBm,inwhichribAandvirB8areco-transcribedandbindcommontranscriptionfactors(LiandCarlow2012),relativeabundancelevelssuggestthatinBwStr,ribAistranscribedindependentlyofthevirB8-D4operon.
SomeWOL-B-strains,suchasBwVulC,lackwspBatthe3′-terminusofthevirB8-D4operon,whileourdataconfirmthatinBwStr,wspBisco-transcribedwiththevirgenes,consistentwithsimilarrelativeabundancesofWspBandthefiveVirproteins.
Inaggregate,theseobser-vationssuggestthatWOL-DandWOL-A-/B-strainsmaydifferinhowRibAandWspBexpressioninterfaceswith66ArchMicrobiol(2016)198:53–69T4SS-mediatedtransportofeffectorsinfilarialwormsandarthropodhosts(Felixetal.
2008;Masuietal.
2000;Rancesetal.
2008;Wuetal.
2004),anditwillbeofinteresttoexplorewhethersuchdifferencesrelatetoriboflavinpro-visioning.
Infilarialnematodes(LiandCarlow2012;Stru-bingetal.
2010;Wuetal.
2009)andbedbugs(Hosokawaetal.
2010),evidencesuggeststhatWolbachiaprovisionshostwithriboflavin,theprecursorofflavincofactorsthatareessentialformanycellularredoxreactions.
Incontrast,riboflavindepletionreducesBwStrabundanceinC/wStr1cells,suggestingthatBwStrutilizeshostriboflavinanddoesnotaugmentriboflavinlevelsinmosquitohostcells(Fallonetal.
2014).
PotentialfunctionsofRibAandRibBIninitialcommitmentstepsinriboflavinbiosynthesis,enzymaticactivitiesencodedbytheribAandribBfunc-tionaldomainsuseGTPandribulose-5-phosphateassub-stratestocatalyzeriboflavinbiosynthesis,consuming25moleculesofATPpermoleculeofriboflavin(Bacheretal.
2000).
WenotethatinWolbachiagenomes,ribAistheannotatedhomologofribBAinEscherichiacoli(Brutineletal.
2013)andencodesadihydroxybutanonephosphatesynthasedomainwithputativeRibBfunctionneartheN-terminus,upstreamofaGTPcyclohydrolaseIIdomainwithconserveddimerizationandactivesiteresidues(RibAfunction).
AsinE.
coli,WolbachiagenomesalsoencoderibB,butatadistinctchromosomallocus,suggestingthatribAandribBarenotcoordinatelyexpressed.
InSinorhizo-biummeliloti(Rhizobiales;Alphaproteobacteria),knock-outmutationsofribBAdecreasedflavinsecretionbutdidnotcauseriboflavinauxotrophyorblockestablishmentofsymbiosis,suggestingthatRibBAmayhaveanundefinedroleinmoleculartransport(Yurgeletal.
2014).
AsisthecasewithBwStr,RibBisatleastthreefoldmoreabundantthanRibAinthebacteriumAcidithiobacillusferrooxidans(Knegtetal.
2008).
Inyeast,RibBhasthiol-dependentalternativeredoxstates(McDonaghetal.
2011),partiallylocalizestothemitochondrialperiplasm,andhasanunex-plainedfunctioninoxidativerespirationthatisindependentofriboflavinbiosynthesis(Jinetal.
2003).
Theseobserva-tionsraisethepossibilitythatinWolbachia,RibAandRibBmayhavefunctionsotherthanriboflavinbiosynthesisthatintegratewithpathwaysinvolvedincellularoxidativestate,suchasironmetabolism.
Intracellularbacteriaarechal-lengedbyhost-imposedoxidativestressandironstarvation(reviewedbyBenjaminetal.
2010)andriboflavinbiosyn-thesisisassociatedwithironacquisitioninbacteriasuchasHelicobacterpylori(Worstetal.
1998)andCampylobacterjejuni(Crossleyetal.
2007).
Wolbachiainterfereswithironmetabolismandsequestrationininsects(Brownlieetal.
2009;Kremeretal.
2009)andinfluencesiron-dependenthostprocessessuchashememetabolism,oxidativestress,apoptosisandautophagy(Gilletal.
2014).
Wenotethattheperiplasmiciron-bindingcomponentofamembranetrans-porterisanabundantproteininBwStr(TableS3,entry778andBaldridgeetal.
2014).
AcknowledgmentsThisworkwassupportedbyGrantAI081322fromtheNationalInstitutesofHealthandbytheUniversityofMin-nesotaAgriculturalExperimentStation,St.
Paul,MN.
CompliancewithethicalstandardsConflictofinterestTheauthorshavenoconflictsofinteresttodeclare.
OpenAccessThisarticleisdistributedunderthetermsoftheCrea-tiveCommonsAttribution4.
0InternationalLicense(http://creativecom-mons.
org/licenses/by/4.
0/),whichpermitsunrestricteduse,distribution,andreproductioninanymedium,providedyougiveappropriatecredittotheoriginalauthor(s)andthesource,providealinktotheCreativeCommonslicense,andindicateifchangesweremade.
ReferencesAlvarez-MartinezCE,ChristiePJ(2009)BiologicaldiversityofprokaryotictypeIVsecretionsystems.
MicrobiolMolBiolRev73:775–808BacherA,EberhardtS,FischerM,KisK,RichterG(2000)Biosyn-thesisofvitaminB2(riboflavin).
AnnuRevNutr20:153–167BaldoL,LoN,WerrenJH(2005)MosaicnatureoftheWolbachiasurfaceprotein.
JBacteriol187:5406–5418.
doi:10.
1128/JB.
187.
15.
5406-5418.
2005BaldoL,BordensteinS,WerengreenJJ,WerrenJH(2006a)Wide-spreadrecombinationthroughoutWolbachiagenomes.
MolBiolEvol23:437–449BaldoL,DunningHotoppJC,JolleyKA,BordensteinSR,BiberSA,ChoudhuryRR,HayashiC,MaidenMC,TettelinH,WerrenJH(2006b)Multilocussequencetypingsystemfortheendosymbi-ontWolbachiapipientis.
ApplEnvironMicrobiol72:7098–7110BaldoL,DesjardinsCA,RussellJA,StahlhutJK,WerrenJH(2010)Acceleratedmicroevolutioninanoutermembraneprotein(OMP)oftheintracellularbacteriaWolbachia.
BMCEvolBiol.
doi:10.
1186/1471-2148-10-48BaldridgeGD,BaldridgeAS,WitthuhnBA,HigginsL,MarkowskiTW,FallonAM(2014)ProteomicprofilingofarobustWol-bachiainfectioninanAedesalbopictusmosquitocellline.
MolMicrobiol94:537–556.
doi:10.
1111/mmi.
12768BearePA,SandozKM,OmslandA,RockeyDD,HeinzenRA(2011)Advancesingeneticmanipulationofobligateintracellularbacte-ria.
FrontMicrobiol2:97.
doi:10.
3389/fmicb.
2011.
00097BenjaminJA,DesnoyersG,MorissetteA,SalvailH,MasséE(2010)Dealingwithoxidativestressandironstarvationinmicroorgan-isms:anoverview.
CanJPhysiolPharmacol88:264–272BennuruS,SemnaniR,MengZ,RibeiroJMC,VeenstraTD,NutmanTB(2009)Brugiamalayiexcreted/secretedproteinsatthehost/parasiteinterface:stage-andgender-specificproteomicprofiling.
PLoSNeglTropDis3:e410.
doi:10.
1371/journal.
pntd.
0000410BigelowHR,PetreyDS,LiuJ,PrzybylskiD,RostB(2004)Predict-ingtransmembranebeta-barrelsinproteomes.
NucleicAcidsRes32:2566–2577BordensteinSR,ReznikoffWS(2005)MobileDNAinobligateintra-cellularbacteria.
NatRevMicrobiol3:688–69967ArchMicrobiol(2016)198:53–69BourtzisK(2008)Wolbachia-basedtechnologiesforinsectpestpopulationcontrol.
AdvExpMedBiol627:104–113.
doi:10.
1007/978-0-387-78225-6_9BraytonKA,PalmerGH,LundgrenA,YiJ,BarbetAF(2002)Anti-genicvariationofAnaplasmamarginalemsp2occursbycombi-natorialgeneconversion.
MolMicrobiol43:1151–1159BrownlieJC,CassBN,RieglerM,WitsenburgJJ,Iturbe-OrmaetxeI,McGrawEA,O'NeillSL(2009)Evidenceformetabolicpro-visioningbyacommoninvertebrateendosymbiont,Wolbachiapipientis,duringperiodsofnutritionalstress.
PLoSPathog5:e1000368.
doi:10.
1371/journal.
ppat.
1000368BrutinelED,DeanAM,GralnickJA(2013)Descriptionofaribofla-vinbiosyntheticgenevariantprevalentinthephylumproteobac-teria.
JBacteriol195:5479–5486ChávezAS,FelsheimRF,KurttiTJ,KuPS,BraytonKA,MunderlohUG(2012)ExpressionpatternsofAnaplasmamarginaleMsp2variantschangeinresponsetogrowthincattle,andtickcellsversusmammaliancells.
PLoSOne7(4):e36012.
doi:10.
1371/journal.
pone.
0036012ComandatoreF,SasseraD,MontagnaM,KumarS,DarbyA,BlaxterMetal(2013)Phylogenomicsandanalysisofsharedgenessug-gestasingletransitiontomutualisminWolbachiaofnematodes.
GenomeBiolEvol5:1668–1674CordauxR,Michel-SalzatA,BouchonD(2001)Wolbachiainfectionincrustaceans:novelhostsandpotentialroutesforhorizontaltransmission.
JEvolBiol14:237–243CordauxR,PichonS,LingA,PerezP,DelaunayC,VavreF,BouchonD,GreveP(2008)Intensetranspositionalactivityofinsertionsequencesinanancientendosymbiont.
MolBiolEvol25:1889–1895.
doi:10.
1093/molbev/msn134CrossleyRA,GaskinDJ,HolmesK,MulhollandF,WellsJM,KellyDJ,vanVlietAH,WaltonNJ(2007)RiboflavinbiosynthesisisassociatedwithassimilatoryferricreductionandironacquisitionbyCampylobacterjejuni.
ApplEnvironMicrobiol73:7819–7825DedeineF,BandiC,BouletreauM,KramerL(2003)InsightsintoWolbachiaobligatorysymbiosis.
In:BourtzisK,MillerTA(eds)Insectsymbiosis.
CRCPress,Florida,pp267–282DellA,GaladariA,SastreF,HitchenP(2010)Similaritiesanddif-ferencesintheglycosylationmechanismsinprokaryotesandeukaryotes.
IntJMicrobiol.
doi:10.
1155/2010/148178FallonAM(2008)CytologicalpropertiesofanAedesalbopictusmos-quitocelllineinfectedwithWolbachiastrainwAlbB.
InVitroCellDevBiolAnim44:154–161.
doi:10.
1007/s11626-008-9090-4FallonAM,BaldridgeGD,HigginsLA,WitthuhnBA(2013)Wol-bachiafromtheplanthopperLaodelphaxstriatellusestablishesarobust,persistent,streptomycin-resistantinfectioninclonalmos-quitocells.
InVitroCellDevBiolAnim49:66–73.
doi:10.
1007/s11626-012-9571-3FallonAM,BaldridgeGD,CarrollEM,KurtzCM(2014)DepletionofhostcellriboflavinreducesWolbachiainculturedmosquitocells.
InVitroCellDevBiolAnim50:707–713.
doi:10.
1007/s11626-014-9758-xFelixC,PichonS,Braquart-VarnierC,BraigH,ChenL,GarrettRA,MartinG,GreveP(2008)CharacterizationandtranscriptionalanalysisoftwogeneclustersfortypeIVsecretionmachin-eryinWolbachiaofArmadillidiumvulgare.
ResMicrobiol159:481–485FloateKD,CoghlinPC,DosdallL(2011)AtestusingWolbachiabacteriatoidentifyEurasiansourcepopulationsofCabbageSeedPodWeevil,Ceutorhynchusobstrictus(Marsham),inNorthAmerica.
EnvironEntomol40:818–2011.
doi:10.
1603/EN10315GillAG,DarbyAC,MakepeaceBL(2014)Ironnecessity:thesecretofWolbachia'ssuccessPLoSNeglTropDis16:e3224.
doi:10.
1371/journal.
pntd.
0003224GillespieJJ,AmmermanNC,Dreher-LesnickSM,RahmanMS,Wor-leyMJ,SetubalJC,SobralBS,AzadAF(2009)AnanomaloustypeIVsecretionsysteminRickettsiaisevolutionarilycon-served.
PLoSOne4:e4833.
doi:10.
1371/journal.
pone.
0004833GillespieJJ,BraytonKA,WilliamsKP,QuevadoDiazMA,BrownWC,AzadAF,SobralBW(2010)PhylogenomicsrevealsadiverseRickettsialestypeIVsecretionsystem.
InfectImmun78:1809–1823HilgenboeckerK,HammersteinP,SchlattmannP,TelschowA,Wer-renJH(2008)HowmanyspeciesareinfectedwithWolbachiaAstatisticalanalysisofcurrentdata.
FEMSMicrobiolLett281:215–220HosokawaT,KogaR,KikuchiY,MengX-Y,FukatsuT(2010)Wol-bachiaasabacteriocyte-associatednutritionalmutualist.
ProcNatlAcadSciUSA107:769–774HotoppJC,LinM,MadupuR,CrabtreeSV,AngilouiSVetal(2006)Comparativegenomicsofemerginghumanehrlichiosisagents.
PLoSGenet2:e21JamesAC,DeanMD,McMahonME,BallardJW(2002)DynamicsofdoubleandsingleWolbachiainfectionsinDrosophilasimu-lansfromNewCaledonia.
Heredity88:182–189JinC,BarrientosA,TzagoloffA(2003)Yeastdihydroxybutanonephosphatesynthase,anenzymeoftheriboflavinbiosyntheticpathway,hasasecondunrelatedfunctioninexpressionofmito-chondrialrespiration.
JBiolChem278:14698–14703KambrisZ,CookPE,PhucHK,SinkinsSP(2009)Immuneactivationbylife-shorteningWolbachiaandreducedfilarialcompetenceinmosquitoes.
Science326:134–136.
doi:10.
1126/science.
1177531KlassonL,WestbergJ,SapountzisP,NaslundK,LutnaesY,DarbyAC,VenetiZ,ChenL,BraigHR,GarretRetal(2009)ThemosaicstructureoftheWolbachiawRistraininfectingDrosoph-ilasimulans.
ProcNatlAcadSciUSA106:5725–5730KnegtFH,MelloLV,ReisFC,SantosMT,VicentiniR,FerrazLF,OttoboniLM(2008)ribBandribBAgenesfromAcidithiobacil-lusferrooxidans:expressionlevelsunderdifferentgrowthcondi-tionsandphylogeneticanalysis.
ResMicrobiol159:423–431KoebnikR,LocherKP,VanGelderP(2000)Structureandfunctionofbacterialoutermembraneproteins:barrelsinanutshell.
MolMicrobiol37:239–253KremerN,VoroninD,CharifD,MavinguiP,MollereauB,VavreF(2009)Wolbachiainterfereswithferritinexpressionandironmetabolismininsects.
PLoSPathog5:e1000630.
doi:10.
1371/journal.
ppat.
1000630LiZ,CarlowCKS(2012)CharacterizationoftranscriptionfactorsthatregulatethetypeIVsecretionsystemandriboflavinbio-synthesisinWolbachiaofBrugiamalayi.
PLoSOne7:e51597.
doi:10.
1371/journal.
pone0051597LiuH,BaoW,LinM,NiuH,RikihisaY(2012)EhrlichiatypeIVsecretioneffectorECH0825istranslocatedtomitochondriaandcurbsROSandapoptosisbyupregulatingMnSOD.
CellMicro-biol14:1037–1050.
doi:10.
1111/j.
1462-5822.
2012.
01775.
xLoN,ParaskevopoulosC,BourtzisK,O'NeillSL,WerrenJH,Bor-densteinSR,BandiC(2007)Taxonomicstatusoftheintracel-lularbacteriumWolbachiapipientis.
IntJSystEvolMicrobiol57:654–657LockwoodS,VothDE,BraytonKA,BearePA,BrownWC,HeinzenRA,BroschatSL(2011)IdentificationofAnaplasmamarginaletypeIVsecretionsystemeffectorproteins.
PLoSOne6:e27724.
doi:10.
1371/journal.
pone.
0027724MasuiS,SasakiT,IshikawaH(2000)GenesforthetypeIVsecre-tionsysteminanintracellularsymbiont,Wolbachia,acausativeagentofvarioussexualalterationsinarthropods.
JBacteriol182:6529–6531McDonaghB,ReguejoR,Fuentes-AlmagroCA,OguetaS,BárcenaJA,PadillaCA(2011)Thiolredoxproteomicsidentifiesdiffer-entialtargetsofcytosolicandmitochondrialglutaredoxin-2iso-formsinSaccharomycescerevisiae.
ReversibleS-glutathionyla-tionofDHBPsynthase(RIB3).
JProteomics74:2487–249768ArchMicrobiol(2016)198:53–69MelnikowE,XuS,LiuJ,LiL,OksovY,GhedinEetal(2011)Inter-actionofaWolbachiaWSP-likeproteinwithanuclear-encodedproteinofBrugiamalayi.
IntJParasitol41:1053–1061MorrowJL,FrommerM,ShearmanDC,RieglerM(2014)Tropi-caltephritidfruitflycommunitywithhighincidenceofsharedWolbachiastrainsasplatformforhorizontaltransmis-sionofendosymbionts.
EnvironMicrobiol16:3622–3637.
doi:10.
1111/1462-2920.
12382NewtonLG,BordensteinSR(2011)Correlationsbetweenbacte-rialecologyandmobileDNA.
CurrMicrobiol62:198–208.
doi:10.
1007/s00284-010-9693-3NiuH,Kozjak-PavlovicV,RudelT,RikihisaY(2010)AnaplasmaphagocytophilumAts-1isimportedintohostcellmitochon-driaandinterfereswithapoptosisinduction.
PLoSPathog6:e1000774NodaH,KoizumiY,ZhangQ,DengK(2001a)InfectiondensityofWolbachiaandincompatibilitylevelintwoplanthopperspecies,LaodelphaxstriatellusandSogatellafurcifera.
InsectBiochemMolBiol31:727–737NodaH,MiyoshiT,ZhangQ,WatanabeK,DengK,HoshizakiS(2001b)Wolbachiainfectionsharedamongplanthoppers(Homoptera:Delphacidae)andtheirendoparasite(Strepsiptera:Elenchidae):aprobablecaseofinterspeciestransmission.
MolEcol10:2101–2106NodaH,MyoshiT,KoizumiY(2002)InvitrocultivationofWol-bachiaininsectandmammaliancelllines.
InVitroCellDevBiolAnim38:423–427NothaftH,SzymanskiCM(2010)Proteinglycosylationinbacteria:sweeterthanever.
NatRev8:775–778.
doi:10.
1038/nrmicro2383OfranY,RostB(2006)ISIS:interactionsitesidentifiedfromsequence.
Bioinformatics23:e13–e16.
doi:10.
1093/bioinformatics/bt303OhashiN,ZhiN,LinQ,RikihisaY(2002)CharacterizationandtranscriptionalanalysisofgeneclustersforatypeIVsecretionmachineryinhumangranulocyticandmonocyticehrlichiosisagents.
InfectImmun70:2128–2138O'NeillSL,PettigrewMM,SinkinsSP,BraigHR,AndreadisTG,TeshRB(1997)InvitrocultivationofWolbachiapipientisinanAedesalbopictuscellline.
InsectMolBiol6:33–39PanX,ZhouG,WuJ,BianG,LuP,RaikhelAS,XiZ(2012)Wol-bachiainducesreactiveoxygenspecies(ROS)-dependentacti-vationofthetollpathwaytocontroldenguevirusinthemos-quitoAedesaegypti.
ProcNatlAcadSciUSA109:E23–E31.
doi:10.
1073/pnas.
1116932108Perrot-MinnotMJ,GuoLR,WerrenJH(1996)SingleanddoubleinfectionswithWolbachiaintheparasiticwaspNasoniavitrip-ennis:effectsoncompatibility.
Genetics143:961–972PichonS,BouchonD,CordauxR,ChenL,GarretRA,GreveP(2009)ConservationofthetypeIVsecretionsystemthrough-outWolbachiaevolution.
BiochemBiophysResCommun385:557–562RancesE,VoroninD,Tran-VanV,MavanguiP(2008)GeneticandfunctionalcharacterizationofthetypeIVsecretionsysteminWolbachia.
JBacteriol190:5020–5030RaychoudhuryR,BaldoL,OliveiraDCSG,WerrenJH(2008)ModesofacquisitionofWolbachia:horizontaltransfer,hybridintro-gression,andconvergenceintheNasoniaspeciescomplex.
Evo-lution63:165–183.
doi:10.
1111/j.
1558-5646.
2008.
00533.
xRioR,HuY,AksoyS(2004)Strategiesofthehome-team:symbiosesexploitedforvector-bornediseasecontrol.
TrendsMicrobiol12:325–336.
doi:10.
1016/j.
tim.
2004.
05.
001SanogoYO,DobsonSL,BordensteinSR,NovakRJ(2007)Disrup-tionoftheWolbachiasurfaceproteingenewspBbyatranspos-ableelementinmosquitoesoftheCulexpipienscomplex(Dip-tera,Culicidae).
InsectMolBiol16:143–154SaridakiA,BourtzisK(2010)Wolbachia:morethanjustabugininsectgenitals.
CurrOpinMicrobiol13:67–72SarkarM,TroeseMJ,KearnsSA,YangT,ReneerDV,CarlyonJA(2008)AnaplasmaphagocytophilumMSP2(P44)-18pre-dominatesandismodifiedintomultipleisoformsinhumanmyeloidcells.
InfectImmun76:2090–2098.
doi:10.
1128/IAI.
01594-07SchulerH,BertheauC,EganSP,FederJL,RieglerM,Schlick-SteinerBC,SteinerFM,JohannesenJ,KernP,TubaK,Laka-tosF,KopplerK,ArthoferW,StaufferC(2013)EvidenceforarecenthorizontaltransmissionandspatialspreadofWolbachiafromendemicRhagoletiscerasi(Diptera:Tephritidae)toinva-siveRhagoletiscingulatainEurope.
MolEcol22:4101–4111.
doi:10.
1111/mec.
12362ShihKM,GerendayA,FallonAM(1998)CultureofmosquitocellsinEagle'smedium.
InVitroCellDevBiolAnim34:629–630SieversF,WilmA,DineenD,GibsonTJ,KarplusK,LiW,LopezR,McWilliamH,RemmertM,SodungJ,ThompsonJD,HigginsDG(2011)Fast,scalablegenerationofhigh-qualityproteinmul-tiplesequencealignmentsusingClustalOmega.
MolSystBiol7:539.
doi:10.
1038/msb.
2011.
75SinkinsSP,GouldF(2006)Genedrivesystemsforinsectdiseasevec-tors.
NatRevGenet7:427–435StrubingU,LuciusR,HoeraufA,PfarrKM(2010)Mitochondrialgenesforheme-dependentrespiratorychaincomplexesareup-regulatedafterdepletionofWolbachiafromfilarialnematodes.
IntJParasitol15:1193–1202.
doi:10.
1016/j.
ijpara.
2010.
03.
004SwoffordDL(2002)PAUP*.
Phylogeneticanalysisusingparsimony(*andothermethods).
Version4.
SinauerAssociates,SunderlandTaylorMJ,BandiC,HoeraufA(2005)Wolbachiabacterialendosym-biontsoffilarialnematodes.
AdvParasitol60:245–284UdayJ,PuttarajuHP(2012)ComparativeanalysisofWolbachiasur-faceproteininD.
melanoagster,A.
tabidaandB.
malayi.
Bioin-formation8(15):711–715.
doi:10.
6026/97320630008711WerrenJH,BaldoL,ClarkME(2008)Wolbachia:mastermanipula-torsofinvertebratebiology.
NatRevMicrobiol6:741–751WorstDJ,GerritsMM,Vandenbroucke-GraulsCM,KustersJG(1998)HelicobacterpyloriribBA-mediatedriboflavinproduc-tionisinvolvedinironacquisition.
JBacteriol180:1473–1479WuM,SunLV,VamathevanJ,RieglerM,DeboyR,BrownlieJC,McGrawE,MartinW,EsserCetal(2004)PhylogenomicsofthereproductiveparasiteWolbachiapipientiswMel:astream-linedgenomeoverrunbymobilegeneticelements.
PLoSBiol2:0327–0341WuB,NovelliJ,FosterJ,VaisvilaR,ConwayL,IngramJ,GanatraM,RaoAU,HamzaI,SlatkoB(2009)ThehemebiosyntheticpathwayoftheobligateWolbachiaendosymbiontofBrugiamalayiasapotentialanti-filarialdrugtarget.
PLoSNeglTropDis3:e475.
doi:10.
1371/journal.
pntd.
0000475YangC-H,XiaoJ-H,NiuL-M,MaG-C,CookJM,BianS-N,FuY-G,HuangD-H(2012)ChaosofWolbachiasequencesinsidethecompactfigsyconiaofFicusbenjamina(Ficus:Moraceae).
PLoSOne.
doi:10.
1371/journal.
pone.
0048882YurgelSN,RiceJ,DomreisE,LynchJ,SaN,QamarZ,RajamaniS,GaoM,RojeS,BauerWD(2014)Sinorhizobiummelilotiflavinsecretionandbacteria-hostinteraction:roleofthebifunc-tionalRibBAprotein.
MolPlant-MicrobeInteract27:437–445.
doi:10.
1094/MPMI-11-13-0338-RZabalouS,RieglerM,TheodorakopoulouM,StaufferC,SavakisC,BourtzisK(2004)Wolbachia-inducedcytoplasmicincompatibil-ityasameansforinsectpestpopulationcontrol.
ProcNatlAcadSciUSA101:15042–15045.
doi:10.
1073/pnas.
0403853101ZechnerEL,LangS,SchildbachJF(2012)Assemblyandmecha-nismsofbacterialtypeIVsecretionmachines.
PhilTransRSocB367:1073–1087.
doi:10.
1098/rstb.
2011.
020769ArchMicrobiol(2016)198:53–69ZhangKJ,HanX,HongXY(2013)VariousinfectionstatusandmolecularevidenceforhorizontaltransmissionandrecombinationofWolbachiaandCardiniumamongriceplanthoppersandrelatedspecies.
InsectSci3:329–344.
doi:10.
1111/j.
1744-7917.
2012.
01537.
xZugR,HammersteinP(2014)BadguysturnedniceAcriticalassessmentofWolbachiamutualismsinarthropodhosts.
BiolRev.
doi:10.
1111/brv.
12098ZugR,KoehnckeA,HammersteinP(2012)Epidemiologyinevo-lutionarytime:thecaseofWolbachiahorizontaltransmis-sionbetweenarthropodspecies.
JEvolBiol25:2149–2160.
doi:10.
1111/j.
1420-9101.
2012.
02601.
x
1007/s00203-015-1154-8ORIGINALPAPERMosaiccompositionofribAandwspBgenesflankingthevirB8D4operonintheWolbachiasupergroupBstrain,wStrGeraldD.
Baldridge1·YangGraceLi1·BruceA.
Witthuhn2·LeeAnnHiggins2·ToddW.
Markowski2·AbigailS.
Baldridge3·AnnM.
Fallon1Received:27April2015/Revised:9September2015/Accepted:14September2015/Publishedonline:23September2015TheAuthor(s)2015.
ThisarticleispublishedwithopenaccessatSpringerlink.
comvirproteinsareexpressedatsimilar,aboveaverageabun-dancelevels.
InwStr,bothribAandwspBaremosaicsofconservedsequencemotifsfromWolbachiasupergroupA-andB-strains,andwspBisnearlyidenticaltoitshomologfromwCobU4-2,anA-strainfromweevils(Coleoptera).
WedescribeconservedrepeatedsequenceelementsthatmapwithinornearpseudogenelesionsandtransitionsbetweenA-andB-strainmotifs.
ThesestudiescontributetoongoingeffortstoexploreinteractionsbetweenWolbachiaanditshostcellinaninvitrosystem.
KeywordsWolbachia·LC–MS/MS·Proteomics·Mosaicgenes·T4SS·RibA·RibB·WspBIntroductionWolbachiapipientis(Rickettsiales;Alphaproteobacteria)isanobligateintracellularbacteriumthatinfectsfilar-ialnematodesandawiderangeofarthropodsincluding≥60%ofinsectsand≈35%ofisopodcrustaceans,butdoesnotinfectvertebrates(Hilgenboeckeretal.
2008).
WolbachiaisconsideredtobeasinglespeciesclassifiedintocladesbymultilocussequencetypinganddesignatedassupergroupsAtoN(Baldoetal.
2006b;Comandatoreetal.
2013;Loetal.
2007).
TheC-andD-strainsthatinfectfilarialwormshavephylogeniesconcordantwiththoseofnematodehosts,consistentwithstrictverticaltransmissionasobligatemutualists(Comandatoreetal.
2013;Dedeineetal.
2003;LiandCarlow2012;Strubingetal.
2010;Tay-loretal.
2005;Wuetal.
2004).
Althougharthropod-asso-ciatedA-andB-strainsmayprovidesubtlefitnessbenefitstohosts(ZugandHammerstein2014),theyarebestknownasreproductiveparasites,causingphenotypesthatmain-tainorincreaseWolbachiainfectionfrequencies,includingAbstractTheobligateintracellularbacterium,Wol-bachiapipientis(Rickettsiales),isawidespread,verti-callytransmittedendosymbiontoffilarialnematodesandarthropods.
Ininsects,Wolbachiamodifiesreproduction,andinmosquitoes,infectioninterfereswithreplicationofarboviruses,bacteriaandplasmodia.
DevelopmentofWol-bachiaasatooltocontrolpestinsectswillbefacilitatedbyanunderstandingofmoleculareventsthatunderliegeneticexchangebetweenWolbachiastrains.
Here,weusednucle-otidesequence,transcriptionalandproteomicanalysestoevaluateexpressionlevelsandestablishthemosaicnatureofgenesflankingtheT4SSvirB8-D4operonfromwStr,asupergroupB-strainfromaplanthopper(Hemiptera)thatmaintainsarobust,persistentinfectioninanAedesalbop-ictusmosquitocellline.
Basedonproteinabundance,ribA,whichcontainspromoterelementsatthe5′-endoftheoperon,isweaklyexpressed.
The3′-endoftheoperonencodesanintactwspB,whichencodesanoutermembraneproteinandisco-transcribedwiththevirgenes.
WspBandCommunicatedbyMarkusNett.
ElectronicsupplementarymaterialTheonlineversionofthisarticle(doi:10.
1007/s00203-015-1154-8)containssupplementarymaterial,whichisavailabletoauthorizedusers.
*AnnM.
Fallonfallo002@umn.
edu1DepartmentofEntomology,UniversityofMinnesota,1980FolwellAve.
,St.
Paul,MN55108,USA2DepartmentofBiochemistry,MolecularBiologyandBiophysics,UniversityofMinnesota,Minneapolis,MN55455,USA3FeinbergSchoolofMedicine,NorthwesternUniversity,Chicago,IL60611,USA54ArchMicrobiol(2016)198:53–69feminization,parthenogenesis,andcytoplasmicincompat-ibility(SaridakiandBourtzis2010;Werrenetal.
2008).
Interferencewithhostimmunemechanismsandreplicationofarboviruses,bacteriaandmalarialplasmodia(Kambrisetal.
2009;Panetal.
2012;ZugandHammerstein2014)hasencouragedeffortstoexploitWolbachiaforbiocontrolofarthropodvectorsofvertebratepathogensand/orcroppests(Bourtzis2008;Rioetal.
2004;SinkinsandGould2006;Zabalouetal.
2004).
AnunderstandingofmoleculardifferencesbetweenA-andB-strains,andhowtheyhavebeeninfluencedbyhorizontaltransmissionandgeneticexchange(NewtonandBordenstein2011;Schuleretal.
2013;Werrenetal.
2008;ZugandHammerstein2014)willfacilitatemanipulationofWolbachia.
Wolbachia'sinteractionwithhostcellslikelyinvolvesthetypeIVsecretionsystem(T4SS),amacromolecularcomplexthattransportsDNA,nucleoproteinsand"effec-tor"proteinsacrossthemicrobialcellenvelopeintothehostcell,wheretheymediateintracellularinteractions(Alvarez-MartinezandChristie2009;Zechneretal.
2012).
HomologsofallgenesexceptvirB5ofAgrobacteriumtumefaciensT4SShavebeenidentifiedinWolbachiaandothermembersoftheRickettsiales(Gillespieetal.
2009,2010),includingAnaplasma,Ehrlichia,Neorickettsia,OrientiaandRickettsia.
AmongsequencedWolbachiagenomes,T4SSgenesareorganizedintwooperons:virB3-B6containingvirB3,virB4andfourvirB6paralogsandvirB8-D4containingvirB8,virB9,virB10,virB11,virD4and,insomegenomes,thewspBparalogofthewspAmajorsurfaceantigen(Pichonetal.
2009;Rancesetal.
2008).
InthesupergroupB-strainwPipfromCulexpipiensmosqui-toes,wspBisdisruptedbyatransposonandispresumablyinactive(Sanogoetal2007).
T4SSeffectorproteinsthatmanipulatehostcellshavebeenidentifiedfromAnaplasmaandEhrlichia(Liuetal.
2012;Lockwoodetal.
2011;Niuetal.
2010),andWolbachiaexpressbothviroperonsinovariesofarthropodhosts,whereinT4SSeffectorsaresuspectedtoplayaroleincytoplasmicincompatibilityandotherreproductivedistortions(Masuietal.
2000;Rancesetal.
2008;Wuetal.
2004).
AlthoughWspAandWspBarelikelycomponentsoftheWolbachiaoutermembrane,theirfunctionsremainunknown.
InthecaseofwBm,WspBisexcreted/secretedintofilarialhostcells(Bennuruetal.
2009)andco-localizeswiththeBm1_46455hostproteinintissuesthatincludeembryonicnuclei(Melnikowetal.
2011).
WspBisthereforeitselfacandidateT4SSeffectorthatmayplayaroleinreproductivemanipulationofthehost.
TheWolbachiastrainwStrinsupergroupBcausesstrongcytoplasmicincompatibilityintheplanthopper,Laodelphaxstriatellus(Nodaetal.
2001a),andinaddi-tionmaintainsarobust,persistentinfectioninaclonalAedesalbopictusmosquitocellline,C/wStr1(Fallonetal.
2013;Nodaetal.
2002).
BecauseinvitrostudieswithwStrprovideadvantagesofscaleandeaseofmanipulationforexploringmechanismsthatmayfacilitatetransformationandgeneticmanipulationofWolbachia,wehaveunder-takenproteomics-basedstudiesthatprovidestrongsupportforexpressionofT4SSmachineryincellculture.
Here,wereportthesequenceofthevirB8-D4operon,includingflankinggenesribA,upstreamofvirB8,andwspBdown-streamofvirD4.
WeshowthatwspBisintact,describepro-teinstructurepredictedfromthededucedWspBsequence,andverifyco-transcriptionofwspBwithupstreamvirgenes.
RelativeabundancelevelsofWspBandtheVirB8-D4proteinsinwStrarewellaboveaverage,whileRibAisamongtheleastabundantofMS-detectedproteins.
InwStr,ribAandwspBaremosaicsofsequencemotifsthataredif-ferentiallyconservedinsupergroupA-(WOL-A)andB-(WOL-B)strains,andtheycontainconserved8-bprepeatelementsthatmaybeassociatedwithgeneticexchange.
Finally,wediscussimplicationsforfunctionalintegrationoftheWolbachiaT4SSwithWspBandwiththeribofla-vinbiosynthesispathwayenzymesGTPcyclohydrolaseII(RibA)anddihydroxybutanonephosphatesynthase(RibB).
MaterialsandmethodsCultivationofcellsAedesalbopictusC7-10andC/wStr1cellsweremaintainedinEagle'sminimalmediumsupplementedwith5%fetalbovineserumat28–30°Cina5%CO2atmosphere(Fal-lonetal.
2013;Shihetal.
1998).
Cellswereharvesteddur-ingexponentialgrowth,underconditionsfavoringmaximalrecoveryofWolbachia(Baldridgeetal.
2014).
Polymerasechainreaction,cloningandDNAsequencingThepolymerasechainreaction(PCR)wasusedtoamplifywStrgenesfromDNAextractspreparedfromWolbachiaenrichedbyfractionationofC/wStr1cellsonsucroseden-sitygradientsandrecoveredfromtheinterfacebetween50and60%sucrose(Baldridgeetal.
2014).
TemplateDNAwasusedtoobtain21PCRproductsusingapanelof31primers(TableS1),GoTaqDNApolymerase(Promega,Madison,WI),andaTechneTC-312cycler(Staffordshire,UK).
Cycleparameterswere:1cycleat94°Cfor2min,35cyclesat94°Cfor35s,53°Cfor35s,72°Cfor1min,followedby1cycleat72°Cfor5min.
Extensiontimewasincreasedto2minforproducts≥1000bp.
PCRproductswereclonedinthepCR4-TOPOvectorwiththeTOPO-TACloningKitforSequencing(LifeTechnologies,GrandIsland,NY),andtwoormorecloneseachweresequenced55ArchMicrobiol(2016)198:53–69attheUniversityofMinnesotaBioMedicalGenomicsCenter.
ReversetranscriptasepolymerasechainreactionTotalRNAwaspurifiedfromA.
albopictusC7-10andC/wStr1cellsusingthePureLinkRNAMiniKit(LifeTechnologies)andtreatedwithDNaseI(RNase-free;LifeTechnologies)followedbyheatinactivation,assuggestedbythemanufacturer.
RT-PCRwasexecutedwithprimersvirD4F1764–1784andwspBR152–172(TableS1)usingtheRNAPCRCoreKit(LifeTechnologies)assuggestedbythemanufacturerwiththeexceptionthatsynthesizedcDNAwastreatedwithDNase-inactivatedRNaseAbeforethefinalPCRreaction.
ThePCRreactionincluded1cycleat95°Cfor4min,35cyclesat95°Cfor35s,56°Cfor40s,72°Cfor40s,followedby1cycleat72°Cfor3min.
Reactionproductswereelectrophoresedon1%agarosegels,cloned,andsequencedasabove.
SequencealignmentsandproteinstructurepredictionDNAandproteinsequencealignmentswereexecutedwiththeClustalOmegaprogram(Sieversetal.
2011).
Align-mentswereeditedbyvisualinspectionandmodifiedinMicrosoftWord.
WspBproteinstructurepredictionswereobtainedusingtoolsavailableatwww.
predictprotein.
org,includingthePROFtmbprogram(Delletal.
2010)forpre-dictionofbacterialtransmembranebetabarrels(Bigelowetal.
2004)andper-residuepredictionofup-strand,down-strand,periplasmicloopandouterlooppositionsofresidues.
ThePROFisisprogram(OfranandRost2006)wasusedtopredictWspBaminoacidresiduesthatarepotentiallyinvolvedinprotein–proteininteractions.
TreeswereproducedusingPAUP*version4(Swofford2002).
AminoacidswerealignedwithClustalW,usingpairwisealignmentparametersof25/0.
5andmultiplealignmentparametersof10/0.
2forgapopeningandgapextension,respectively.
TheproteinweightmatrixwassettoGonnet.
ThealignmentwassavedasanexusfileandloadedintoPAUP*,andthetreeswerecreatedusingaheuristicsearchwiththecriterionsettoparsimony.
Bootstrap50%major-ity-ruleconsensustreesarebasedon1000replicates,withwBm(WOL-D)astheoutgroup.
Massspectrometry,peptidedetection,proteinidentificationandstatisticalanalysisMassspectrometrydata,generatedusingLC–MS/MSonLTQandOrbitrapVelosmassspectrometersasfourdatasets,weredescribedpreviously(Baldridgeetal.
2014).
TheMSsearchdatabasewasmodifiedtoincludededucedORFsfromwStrsequencedatadescribedherein.
AlltestsofassociationwereperformedwithSASversion9.
3(Cary,NC;http://www.
sas.
com/en_us/home.
html/).
ResultsStructureofthewStrvirB4D8operonTherobust,persistentinfectionofA.
albopictusmos-quitocellline,C/wStr1withBwStr(inthetextbelow,straindesignationsaredenotedbysuperscripts),isolatedfromtheplanthopperL.
striatellus,providesaninvitromodeltoidentifyproteinsthatmodulatethehost–microbeTable1MS-detectedpeptidesfromwStrproteinsencodedbyribA,ribBandthevirB8-D4operonaProteinmassinkilodaltons.
bNumberof95%confidenceuniquepeptides;(1)designatesoriginalsearch[7];(2)designatesarefinedsearchinwhichthedatabaseincludedpeptidesbasedonthepresentwStrnucleotidesequencedata;(T)combinedtotalpeptidesfrombothsearches.
cPercentproteinsequencecoveragerepresentedbydetectedpeptides.
dMeannumberofpeptidesfromfourindependentMSdatasets.
eStudentizedresidualbasedonthemodifiedunivariablemodeloftherefinedsearch(TableS3,col-umnR);SRvalue0indicatesaverageabundanceprotein,0–1aboveaverage,1–2abundantand>2highlyabundant.
Valuesbelow0indicatelowerthanaverageabundance.
fA94%confidencepeptideindicatedinFig.
1Adidnotmeetthethresholdforproteomeinclusionintheoriginalsearch.
ForVirB10,oneoriginallydetectedpeptidewasabsentfromtherefinedsearchProteinakDabPep(1)bPep(2)bPep(T)cCov.
dRALeSRRibA4122260.
52.
30RibB2471212897.
01.
20VirB82691010585.
00.
59VirB93110810456.
20.
84VirB1054141618538.
80.
94VirB1137121414427.
00.
82VirD477121414266.
20.
45WspB312f11507.
21.
0856ArchMicrobiol(2016)198:53–69interaction.
ApotentialrolefortheT4SSissupportedbystrongrepresentationofpeptidesfromVirB8,VirB9,VirB10,VirB11,VirD4(Table1)andassociatedproteinsintheBwStrproteome(Baldridgeetal.
2014).
Despiteitsemergenceasausefulstrainthatgrowswellinvitro,theBwStrgenomeisnotyetavailable.
InWolbachiastrainsforwhichgenomeannotationisavailable,geneorderwithinthevirB8-D4operonisconserved.
Basedontranscriptionalanalysesintherelatedgenera,AnaplasmaandEhrlichia(Pichonetal.
2009),thepromoterlikelymapswithinthe3′-endofribAextendingintotheintergenicspacer(Fig.
1a,blackhorizontalarrowatleft)andisfollowedbyfivecon-secutivevirgenes(Fig.
1b).
InBwPipfromCulexpipiensmosquitoes,wspBisdisruptedbyinsertionofanIS256elementthatencodesatransposaseontheoppositestrand(Fig.
1a,atright;Sanogoetal.
2007).
BecauseVirB8-D4proteinswerehighlysimilartohomologsfromBwPip(Baldridgeetal.
2014),weevaluatedwspBinBwStranditspotentialexpressionasavirB8-D4operonmember,asisthecaseinAwMelandAwRifromDrosophilaspp.
andAwAtab3fromthewaspAsobaratabida(Rancesetal.
2008;Wuetal.
2004).
Intheoriginalproteomicanalysis,threeWspBpeptides(Fig.
1a,tallblackandgrayarrowsrepresent95and94%confidencepeptides,respectively)mappedproximalanddistaltothetransposoninsertioninBwPip,whiletheabsenceofpeptidescorrespondingtothetransposonsuggestedthatwspBisintactinBwStr.
NucleotideanddeducedaminoacidsequencecomparisonsToexaminethevirB4-D4operoninBwStr,wesequencedoverlappingPCRproductsfrom20primerpairs(TableS1)spanning9.
1kbbeginning43bpdownstreamofthe5′-endofribAinotherWolbachiastrainsandendingwithintopAencodedimmediatelydownstreamoftheoperonontheoppositestrand(Fig.
1b,c).
WiththenotableexceptionoftheBwPiptransposon,thenucleotidesequencealignedmost**wMelwPippromoterB8BB10DD4wspBtopAtransposaseribAribB10kb5kbBwVitBwPipwVulCwMelwBmwRiB9B110CDWOL-BBBAADRT-PCRAFig.
1SchematicmapoftheWolbachiaT4SSvirB8-D4operonandcloningstrategyfortheribAtotopAsequencefromBwStr.
aLeftexpandedviewoftheBwStrribAORFdepictedasanarrowshowingthedirectionoftranscription.
Blackhorizontalarrowindicatesaputa-tivepromoterthatextendsintoanintergenicspacer(blackrectangle).
BlackarrowheadsindicatepositionsofMS-detecteduniquepeptides(95%confidence).
Gradientshadingfromwhitetoblackdesignates5′-sequenceidentityresemblingWOL-Atransitioningto3′-sequencemorecloselyresemblingWOL-B-strains.
aRightexpandedviewoftheinterruptedwspBhomologinBwPip.
Blackellipsesindicateposi-tionsofIS256invertedrepeatelementsflankinga1.
2-kbinsertionencodingaMULEdomainsuperfamilytransposase(gi|190571636;pfam10551)ontheoppositestrand(indicatedbythedirectionoftheopenarrow);flankinggrayshadingindicateswspB.
Tallverticalblackandgrayarrowheadsindicatepositionsofuniquepeptides(95and94%confidence,respectively)identifiedintheoriginalMSdatasearch.
Smallgrayarrowsindicate95%confidencepeptidesmatchedinarefineddataset(includingtheBwStrsequencedescribedhere)thatareconservedinWOL-B-strains,andopenarrowheadswithstarsindicatepeptidesuniquetoBwStr.
bSchematicdepictionoftheWol-bachiavirB8-D4operonandflankinggeneswitharrowsdesignat-ingthedirectionoftranscription.
Virgenesaredesignatedinwhitefontonablackbackground;blacksquaresindicateintergenicspac-ers.
GradientshadingindicatesmosaicstructureofanintactwspBinBwStr.
cFilledlinesabovethe10-kbscalemarkerrepresentclonedPCRamplificationproducts(seeTableS1forprimers)thatweresequencedandassembledintotheBwStrribBandribA–topAconsen-sussequence.
ThedoubleslashsymbolsatleftindicatethatribBisnotcontiguouswithdownstreamgenes.
TheopenboxindicatestheRT-PCRamplificationproductfromFig.
2.
dBLASTnalignmentofthe9133-bpBwStrribA–topAsequencetocorrespondingsequencesinBwVitBBwPip,BwVulC,AwRi,AwMelandDwBmgenomes.
Darkfilledlinesindicatesequenceidentity>70%;lightlinesindicatelowsequenceidentity,andtheopenspaceinBwPiprepresentsanalign-mentgap57ArchMicrobiol(2016)198:53–69closelytohomologoussequencesfromBwVitBandBwPip.
Inaddition,wenotedvariabilityinan~0.
3-kbregionofvirB10inBwStrthatwasconservedinBwVitB,BwPipandAwRi,butnotinBwVulC,AwMelandDwBm(Fig.
1d;seeTableS2forGenBankAccessions).
PairwisesequencecomparisonsofthevirB8-D4operonfromBwStrtohomologsfromWolbachiasupergroupA,B,C,DandFstrains(Table2)confirmthatvirB10,withnucleotideidentitiesrangingfrom74–99%,istheleastconservedofthefivevirgenes,andwenotethatKlassonetal.
(2009)attributeddiver-genceofvirB10inAwMelandAwRitogeneticexchangewithaWOL-B-strain.
Collectivelyandasindividuals,thevirgenesfromBwStrhavethehighestnucleotideidentities(~99%)withBwVitBandBwPip.
IdentitieswithfiveA-strainsarelower(range87–91%),loweryet(range80–89%)withtheF-strain,FwCleandfalltoarangeof74–88%withthreenematode-associatedstrains,DwBm,CwOoandCwOv.
Atthe5′-endoftheoperon,ribAwasdistinct,withapproximatelyequiva-lentnucleotideidentitywithhomologsfromA-andB-strains(range91–94%),whilethepartialsequenceoftopAdownstreamoftheoperonhadaconservationpatternsimilartothatofthevirgenes.
Insomecom-parisons,virB8,virB11,virD4andtopAaminoacididentitiesexceednucleotideidentities.
AlthoughribBisnotphysicallyadjacenttothevirB8-D4operoninanno-tatedWolbachiagenomes,ribBfromBwStrismostsim-ilartohomologsfromBwNo(97%nucleotideidentity)andAwMel(90%),butwasexceptionalbecauseidenti-tieswiththreeotherinsect-associatedA-andB-strains(~80%)werelowerthanwithF-,C-andD-strains(range85–87%).
Consistentwithearlierproteomicdata(Baldridgeetal.
2014),inallcomparisonsthatdiscriminatebetweenA-andB-strains,BwStrresem-bledWOL-B,whilevariabilityinribAandwspBflank-ingthevirB8-D4genesexceededthatofthevirgenesthemselves.
Table2PairwisenucleotideandaminoacidcomparisonsWolbachiastrainsfromsupergroupsA,B,C,DandFareindicatedbysuperscripts,withpercentagesofnucleotide(N)andaminoacid(AA)sequenceidentitiestoBwStr.
Dashesindicatesequencesnotavailable,andxxindicatespseudogenes;GenBankAccessionnumbersaregiveninTableS2aPartialgeneandproteinsequences:ribA1040bp,ribB592bp;topA825bp.
Hostassociations:wPip,Culexpipiens—mosquito;wVitB,Nasoniavitripennis—wasp;wTai,Teleogryllustaiwanensis—cricket;wVulC,Armadillidiumvulgare—isopod;wMel,wRi,wAna,wNo,Drosophilaspp.
—fruitfly;wKue,Ephestiakuehniella—moth;wAtab3Asobaratabida—wasp;wBm,wOoandwOvfromfilarialnematodesBrugiamalayi,OnchocercaochengiandO.
volvulus,respectively.
Inthecomparison,valuesof97%orgreaterareshowninitalicsGeneBwPiPBwVitBBwNoBwTaiBwVulCAwMelAwRiNAANAANAANAANAANAANAAribAa9489948993889490939293919289virB89910099100999999100949488868887virB99999999898979797949391899189virB109999999890869896887487748885virB119999979996989799909389958995virD49999999999999999949789928993wspB56xx98968568––––85708570topAa99100991009999––––88878786ribBa8180––9796––––90917978GeneAwAnaAwKueAwAtab3FwCleDwBmCwOoCwOvNAANAANAANAANAANAANAAribAa91889391––8481838082748275virB88887888688888583858183818482virB99189918991898484848482768176virB108884877487738071847076647464virB118995899589958995889486898789virD48993899388928792878786918894wspB83688570857072xx736172497149topAa8685––––8892868884838488ribBa8088––––8787868785xx85xx58ArchMicrobiol(2016)198:53–69ExpressionandrelativeabundancesoftheBwStrvirB4D8proteinsTorefineanearlieroriginalproteomicanalysis(Baldridgeetal.
2014),weincorporatedthePCR-amplifiedBwStrsequencesdescribedheretothedatabaseforpeptideiden-tification[Table1,seecolumnlabeledPep(2)].
Statisti-calanalysisindicatedthatinaunivariablemodel,proteinmolecularweightwasweakly(r2=0.
2221)butsignifi-cantly(p1.
0)foranabundantproteinandroughlyequivalenttoSRval-ues(range1–1.
17)ofhousekeepingproteinssuchasisoci-tratedehydrogenase,ftsZ,ATPsynthaseF0F1αsubunit,andribosomalproteinsS2,S9,L3,L7/L12andL14(TableS3).
Incomparison,WspAwithanSRof2.
17(TableS3,entry63)rankedashighlyabundant,andthemostabundantproteinintheproteomewastheGroELchaperone(entry586),withanSRof3.
66.
ReversetranscriptasePCRconfirmscotranscriptionofwspBwithvirgenesSimilarSRvaluesforWspB,relativetoVirB8-D4,wereconsistentwithevidencethatwspBisco-transcribedwithvirB8-D4inAwMel,AwRiandAwAtab3(Rancesetal.
2008;Wuetal.
2004).
WeusedRT-PCRwithRNAtem-plateverifiedbyPCRtobefreeofDNAcontamination(Fig.
2b,lanes2and3)toamplifya528-bpproductthatwasproducedinreactionscontainingRNAfromC/wStr1cells(Fig.
2a,lane4),butnotinnegativecontrolreac-tions(lanes1and2)orthosewithRNAfromC7-10cells(lane3).
ItssequencematchedtheexpectedBwStrgenomicsequence(Fig.
1c,RT-PCRboxatright),confirmingthatinBwStr,wspBisamemberofthevirB8-D4operon.
InBwStr,ribAisamosaicofconservedWOLAandWOLBsequencemotifsTheribAnucleotidesequencehasbeenshowntocontainregulatoryelementsforexpressionoftheT4SSoperoninAnaplasmaandEhrlichia(Ohashietal.
2002;Pichonetal.
2009).
IncontrasttohighesthomologiesofBwStrvirB8-D4genestoWOL-B-strains,ribAsequenceidentitiesshowedlittledifferencebetweenWOL-Aand-Bhomologs(Table2),butthetwoMS-detectedpeptidescorrespondedtoAwMelandBwPiphomologs,respectively(Fig.
1a).
Alignmentofaminoacidsfrom10RibAhomologs(Fig.
3;WOL-AandWOL-B-strainsareidentifiedatleftinredandblue,respectively)suggestedthatBwStrRibAisatwo-partmosaic,eachcontainingaproteinfunctionaldomain.
Theaminoterminal150residuesinBwStrRibA(Fig.
3)includeashortdihydroxybutanonephosphatesynthasedomainandthefirstdetectedpeptide(residues94–104).
ThisportionofBwStrRibAmatchedsequencesfromthefourA-strainsandasingleB-strain,BwVulC,at29of36variableaminoacids(showninred),whileonlythree(4,39and168inblue)matchedtheotherthreeB-strainsandfour(ingreen)wereunique.
Incontrast,theC-terminal151–347residues,encompassingthesecondpeptide(residues250–258)withinaGTPcyclohydrolasedomain,includedasingleaminoaciduniquetoBwStr,while23(inblue)uni-formlymatchedB-strainsexceptBwVulC,whichcontinuedtoresembletheA-strainsuntilresidue239.
AmongthefourA-strains,theBwRihomologismostsimilarthroughoutthealignmenttotheB-strains,butwithinresidues129–150immediatelyprecedingthecyclohydrolasedomain,itcloselymatchedBwTai,BwPipandBwVitB,whileBwStrABFig.
2ReversetranscriptasePCR(RT-PCR)analysisshowsco-transcriptionofwspBwithvirD4.
aLanes1and2RT-PCRnegativecontrolswithnoRNAorwithnoreversetranscriptase,respectively.
Lanes3and4RT-PCRofRNAfromuninfectedC7-10andinfectedC/wStr1cells,respectively,withvirD4forwardandwspBreverseprimers.
Lane5RT-PCRpositivecontrolwithC/wStr1RNAandWolbachiaprimersS12F/S7R,whichamplifyportionsofaribosomalproteinoperondescribedpreviously(Fallon2008).
bLane1PCRnegativecontrolwithnoTaqenzyme.
Lanes2and3negativecontrollackingRT,withRNAfromuninfectedC7-10andinfectedC/wStr1cells,respectively59ArchMicrobiol(2016)198:53–69Fig.
3AminoacidsequencealignmentofRibAhomologsfromBwStrandWolbachiasupergroupsA(red),B(blue)andD(black)respectively.
Asterisksbelowthealignmentindicateuniversallyconservedresidues.
Uniqueresiduesareingreenfont.
ResiduesconservedinBwStrandamajorityofB-strainsareindarkblue,boldfont,whilethoseindarkred,boldfontareconservedwithamajorityofA-strains.
Residuesconservedintwotofourstrainsareinlightblue,orangeororangeboldfont.
Residueshighlightedingraycorrespondto95%confidencepeptidesdetectedbyLC–MS/MS.
Thedihydroxybutanonephosphatesynthase(RibB)andGTPcyclohydrolaseIIdomains(RibA)areindicatedabovethealignmentwithingreaterthanlessthansymbols.
BoldunderlinedresiduesinAwMelandBwStrindicateconservedactivesiteaminoacids,includ-ingcriticalcysteineresidues.
Doubleunderlinedresiduesindicateaminoacidsinvolvedinthedimerizationinterface.
SeeTables2andS2forhostassociationsandGenBankAccessions.
ThePCR-amplifiedBwStrsequencedoesnotencodetheN-terminalaminoacids;position1correspondstothe15thaminoacid1>DHBPsynthasedomainRibAGTPcyclohydrolaseII180wKueNNHEMRNWCEKNDVIALDTSFINNFQENQDVYEVCKTSLFLKQTQEVNIISYRTESGGREwMelNNHEMRNWCEKNDVIALDTSFINNFQENQDVYEVCKTSLFLKQTQEVNIISYRTESGGREwHaNNHEMRNWCEKNDVIALDTSFINNFQENQDVYEVCKTSLFLKQTQEVNIISYRTESGGREwRiNKYEMRNWCEENDIIALDTLLVNDFQQNQSVYEVCKTSLFLKQTQEVDIISYRTESGGREwVulCNNHEMQNWCEKNDVIALDTSFINNFQENQDVYEVCKTSLFLKQTQEVDIISYRTESGGREwStrNNHEMRNWCEKNDVIALDKSFINNFQENQDVYEVCKTSLFLKQTQEVDIISYRTKSGGREwTaiNKHEMRNWCEENDIIALNTLLVNDFQRNHSVYEVCKTSLFLKQTQEVDIISYRTKSGGREwPipNKHEMRNWCEENDIIALNTLLVNDFQQNHSVYEVCKTSLFLKQTQEVDIISYRTKSGGREwVitBNKHEMRNWCEENDIIALNTLLVNDFQQNHSVYEVCKTSLFLKQTQEVDIISYRTKSGGREwBmDEYEMRGWCEKSDVIALDVLFINNFQQNQDIYEVCKTPLFLKQTQKVNIISYRTCNGRKE181>RibAGTPcyclohydrolaseIIdomainRibAGTPcyclohydrolaseIIdomain300wKueDGRGIGLTNKLRAYSMQRGHNLDTVDANRILGFEDDERSFAVAAKMLKKLNINKIQLLTNwMelDGRGIGLTNKLRAYSMQRGHNLDTVDANRILGFEDDERSFAVAAKMLKKLNINKIQLLTNwHaDGRGIGLTNKLRAYSMQREHNLDTVDANRILGFEDDERSFVVAAKMLKKLNINKIQLLTNwRiDGRGIGLTNKLRAYSVQREHNLDTVDANRILGFEDDERSFVVAAKMLKKLNINKIQLLTNwVulCDGRGIGLANKLRAYSMQRRHNLDTVDANRVLGFEDDERSFAVAVEILKKLDIKKIQLLTNwStrDGRGIGLTNKLRAYSMQRKYNLDTVDANRVLGFEDDERSFAVAAKILKKLNINKIQLLTNwTaiDGRGIGLTNKLRAYSMQRKYNLDTVDANRVLGFEDDERSFAVAAKILKKLNINKIQLLTNwPipDGRGIGLTNKLRAYSMQRKYNLDTVDANRVLGFEDDERSFAVAAKILKKLNINKIQLLTNwVitBDGRGIGLTNKLRAYSMQRKYNLDTVDANRVLGFEDDERSFAVAAKILKKLNINKIQLLKNwBmDGRGIGLTNKLRAYDMQRKYNLDTVDANRILGFEDDERSFAVAAEMLKKLGIKKIQLLTN201>RibAGTPcyclohydrolaseIIdomain98%nucleotideidentityFig.
S2)isconsistentwithexchangeofanapparentlyintactgenebetweenmembersofdistinctWolbachiasupergroupsbyamechanismthatrequiresfurtherinvestigation.
Inten-siveanalysisofthewspAparalogdemonstratesthatintra-genicrecombinationbreakpointsareconcentratedincon-servedregionsoutsideoftheHVRs(Baldoetal.
2005,2010).
CAARTARYrepeatsarenotpresentinwspA,andinwspB,theyoccuronlywithinanddirectlyadjacenttoHVR2atpositionsthatcorrespondtopseudogenelesionsinAwCobU4-2andinBwPip(duetoatranspositioneventinBwPip;Sanogoetal.
2007).
Furthermore,Pichonetal.
(2009)suggestedthattranspositioneventsmayexplainabsenceofwspBinthevirB8-D4operonsofmanyWOL-B-strains.
Inapracticalsense,CAARTARYrepeatsatwspBpseudogenelesionsandWOL-A/Bsequencemotiftransitions(Figs.
S1,S2,S3)suggesttheirinvolvementingeneticexchange.
BecausetransformationofWolbachiahasnotyetbeenachieved,engineeringofCAARTARYrepeatsintovectorsusedsuccessfullytointroduceselecta-blemarkersintoothermembersoftheRickettsiales(seeBeareetal.
2011)meritsinvestigation.
PotentialfunctionsofWspBAlthoughbacterialoutermembraneproteinsareimpor-tantmediatorsofinteractionswithhostcellsandspecificfunction(s)ofbothWspAandWspBremaintobeidenti-fied,theymayhaveuniquefunctionsasporinproteinsinWolbachia,whichlackcellwalls.
ThevirB8-D4operonsofWolbachiaanditssistergenera,AnaplasmaandEhrlichia,aresimilarlyorganized(Gillespieetal.
2010;Hotoppetal.
2006)with3′-terminalgenesencodingmajorsurfacepro-teinsthat,analogoustowspB,areco-transcribedwiththevirgenes(Ohashietal.
2002).
InA.
marginale,afamilyofmsp2pseudogenesundergo"combinatorialgenecon-version"attheexpressionsite(Braytonetal.
2002)andMSP2variantschangeduringgrowthindifferenthostcelltypes,whichlikelyreflectsaresponsetohostimmunitymechanisms(Chávezetal.
2012).
Similarly,Baldoetal.
(2010)proposedthatchangesinWspAHVRregionsplayaroleinhostadaptationandinnateimmunityinteractions,consistentwithvariationinthehigher-orderstructureoftheproteinindifferenthosts(UdayandPuttaraju2012).
HVRsequencechangesinthewspBparalogmayreflectasimilardynamic.
AdditionalevidenceindicatesthatMSP2proteinsareglycosylated(Sarkaretal.
2008),whichisnowanestablishedprocessinpost-translationalmodifica-tioninbacteria(Delletal.
2010;NothaftandSzymanski2010),andwenotethatWspBcontainspotentialglycosyla-tionsites.
AlthoughaninactivatedpseudogeneorabsenceofwspBinvirB8-D4operonsofsomeWolbachiastrainsindicatesthatitisnotabsolutelyrequiredforsurvival,asecretomeanalysisofBrugiamalayishowedthatWspBfromDwBmisexcreted/secretedintofilarialhostcells(Bennuruetal.
2009).
Furthermore,itco-localizeswiththeBm1_46455hostproteinintissuesthatincludeembryonicnuclei(Melnikowetal.
2011).
WspBisthereforeitselfacandidateT4SSeffectorthatmayplayaroleinreproduc-tivemanipulationofthehost.
MosaicisminwspBanditshighrateofevolution(Comandatoreetal.
2013)maythusreflectgeneticchangesthatoptimizeadaptationtoparticu-larhostcellssuchasthoseinreproductivetissuesandfacil-itateexploitationofnewarthropodnichesbyWolbachia.
GeneticplasticityofribAinthevirB8D4operonAsidefromwspBatthe3′-endoftheT4SSvirB8-D4operon,ribAexhibitsgeneticplasticityatits5′-end.
InbothBwStrandBwVulC,ribAisatwo-partmosaicofN-termi-nalWOL-AandC-terminalWOL-Bmotifs.
Incontrast,theinternalvirB8-D4geneshavetypicalB-strainidenti-ties,andinsomestraincomparisons,aminoacididentitiesslightlyexceednucleotideidentities,whichPichonetal.
(2009)attributetostrongselectionagainstnon-synony-mouscodonsubstitutions.
AmongtheinternalvirB8-D4genes,however,Klassonetal.
(2009)suggestthatinAwRi,anespeciallyvariableregioninvirB10islikelyderivedfromgeneticexchangewithaB-strain.
WenoteherethatribAfromAwRicloselyresemblesB-strainhomologswithinavariableregionthatimmediatelyprecedestheGTPcyclohydrolasedomain,whereitshomologinBwStrtran-sitionsfromWOL-AtoWOL-Bsequencemotifs(Fig.
S1,positions387–450).
IncontrasttoDwBm,inwhichribAandvirB8areco-transcribedandbindcommontranscriptionfactors(LiandCarlow2012),relativeabundancelevelssuggestthatinBwStr,ribAistranscribedindependentlyofthevirB8-D4operon.
SomeWOL-B-strains,suchasBwVulC,lackwspBatthe3′-terminusofthevirB8-D4operon,whileourdataconfirmthatinBwStr,wspBisco-transcribedwiththevirgenes,consistentwithsimilarrelativeabundancesofWspBandthefiveVirproteins.
Inaggregate,theseobser-vationssuggestthatWOL-DandWOL-A-/B-strainsmaydifferinhowRibAandWspBexpressioninterfaceswith66ArchMicrobiol(2016)198:53–69T4SS-mediatedtransportofeffectorsinfilarialwormsandarthropodhosts(Felixetal.
2008;Masuietal.
2000;Rancesetal.
2008;Wuetal.
2004),anditwillbeofinteresttoexplorewhethersuchdifferencesrelatetoriboflavinpro-visioning.
Infilarialnematodes(LiandCarlow2012;Stru-bingetal.
2010;Wuetal.
2009)andbedbugs(Hosokawaetal.
2010),evidencesuggeststhatWolbachiaprovisionshostwithriboflavin,theprecursorofflavincofactorsthatareessentialformanycellularredoxreactions.
Incontrast,riboflavindepletionreducesBwStrabundanceinC/wStr1cells,suggestingthatBwStrutilizeshostriboflavinanddoesnotaugmentriboflavinlevelsinmosquitohostcells(Fallonetal.
2014).
PotentialfunctionsofRibAandRibBIninitialcommitmentstepsinriboflavinbiosynthesis,enzymaticactivitiesencodedbytheribAandribBfunc-tionaldomainsuseGTPandribulose-5-phosphateassub-stratestocatalyzeriboflavinbiosynthesis,consuming25moleculesofATPpermoleculeofriboflavin(Bacheretal.
2000).
WenotethatinWolbachiagenomes,ribAistheannotatedhomologofribBAinEscherichiacoli(Brutineletal.
2013)andencodesadihydroxybutanonephosphatesynthasedomainwithputativeRibBfunctionneartheN-terminus,upstreamofaGTPcyclohydrolaseIIdomainwithconserveddimerizationandactivesiteresidues(RibAfunction).
AsinE.
coli,WolbachiagenomesalsoencoderibB,butatadistinctchromosomallocus,suggestingthatribAandribBarenotcoordinatelyexpressed.
InSinorhizo-biummeliloti(Rhizobiales;Alphaproteobacteria),knock-outmutationsofribBAdecreasedflavinsecretionbutdidnotcauseriboflavinauxotrophyorblockestablishmentofsymbiosis,suggestingthatRibBAmayhaveanundefinedroleinmoleculartransport(Yurgeletal.
2014).
AsisthecasewithBwStr,RibBisatleastthreefoldmoreabundantthanRibAinthebacteriumAcidithiobacillusferrooxidans(Knegtetal.
2008).
Inyeast,RibBhasthiol-dependentalternativeredoxstates(McDonaghetal.
2011),partiallylocalizestothemitochondrialperiplasm,andhasanunex-plainedfunctioninoxidativerespirationthatisindependentofriboflavinbiosynthesis(Jinetal.
2003).
Theseobserva-tionsraisethepossibilitythatinWolbachia,RibAandRibBmayhavefunctionsotherthanriboflavinbiosynthesisthatintegratewithpathwaysinvolvedincellularoxidativestate,suchasironmetabolism.
Intracellularbacteriaarechal-lengedbyhost-imposedoxidativestressandironstarvation(reviewedbyBenjaminetal.
2010)andriboflavinbiosyn-thesisisassociatedwithironacquisitioninbacteriasuchasHelicobacterpylori(Worstetal.
1998)andCampylobacterjejuni(Crossleyetal.
2007).
Wolbachiainterfereswithironmetabolismandsequestrationininsects(Brownlieetal.
2009;Kremeretal.
2009)andinfluencesiron-dependenthostprocessessuchashememetabolism,oxidativestress,apoptosisandautophagy(Gilletal.
2014).
Wenotethattheperiplasmiciron-bindingcomponentofamembranetrans-porterisanabundantproteininBwStr(TableS3,entry778andBaldridgeetal.
2014).
AcknowledgmentsThisworkwassupportedbyGrantAI081322fromtheNationalInstitutesofHealthandbytheUniversityofMin-nesotaAgriculturalExperimentStation,St.
Paul,MN.
CompliancewithethicalstandardsConflictofinterestTheauthorshavenoconflictsofinteresttodeclare.
OpenAccessThisarticleisdistributedunderthetermsoftheCrea-tiveCommonsAttribution4.
0InternationalLicense(http://creativecom-mons.
org/licenses/by/4.
0/),whichpermitsunrestricteduse,distribution,andreproductioninanymedium,providedyougiveappropriatecredittotheoriginalauthor(s)andthesource,providealinktotheCreativeCommonslicense,andindicateifchangesweremade.
ReferencesAlvarez-MartinezCE,ChristiePJ(2009)BiologicaldiversityofprokaryotictypeIVsecretionsystems.
MicrobiolMolBiolRev73:775–808BacherA,EberhardtS,FischerM,KisK,RichterG(2000)Biosyn-thesisofvitaminB2(riboflavin).
AnnuRevNutr20:153–167BaldoL,LoN,WerrenJH(2005)MosaicnatureoftheWolbachiasurfaceprotein.
JBacteriol187:5406–5418.
doi:10.
1128/JB.
187.
15.
5406-5418.
2005BaldoL,BordensteinS,WerengreenJJ,WerrenJH(2006a)Wide-spreadrecombinationthroughoutWolbachiagenomes.
MolBiolEvol23:437–449BaldoL,DunningHotoppJC,JolleyKA,BordensteinSR,BiberSA,ChoudhuryRR,HayashiC,MaidenMC,TettelinH,WerrenJH(2006b)Multilocussequencetypingsystemfortheendosymbi-ontWolbachiapipientis.
ApplEnvironMicrobiol72:7098–7110BaldoL,DesjardinsCA,RussellJA,StahlhutJK,WerrenJH(2010)Acceleratedmicroevolutioninanoutermembraneprotein(OMP)oftheintracellularbacteriaWolbachia.
BMCEvolBiol.
doi:10.
1186/1471-2148-10-48BaldridgeGD,BaldridgeAS,WitthuhnBA,HigginsL,MarkowskiTW,FallonAM(2014)ProteomicprofilingofarobustWol-bachiainfectioninanAedesalbopictusmosquitocellline.
MolMicrobiol94:537–556.
doi:10.
1111/mmi.
12768BearePA,SandozKM,OmslandA,RockeyDD,HeinzenRA(2011)Advancesingeneticmanipulationofobligateintracellularbacte-ria.
FrontMicrobiol2:97.
doi:10.
3389/fmicb.
2011.
00097BenjaminJA,DesnoyersG,MorissetteA,SalvailH,MasséE(2010)Dealingwithoxidativestressandironstarvationinmicroorgan-isms:anoverview.
CanJPhysiolPharmacol88:264–272BennuruS,SemnaniR,MengZ,RibeiroJMC,VeenstraTD,NutmanTB(2009)Brugiamalayiexcreted/secretedproteinsatthehost/parasiteinterface:stage-andgender-specificproteomicprofiling.
PLoSNeglTropDis3:e410.
doi:10.
1371/journal.
pntd.
0000410BigelowHR,PetreyDS,LiuJ,PrzybylskiD,RostB(2004)Predict-ingtransmembranebeta-barrelsinproteomes.
NucleicAcidsRes32:2566–2577BordensteinSR,ReznikoffWS(2005)MobileDNAinobligateintra-cellularbacteria.
NatRevMicrobiol3:688–69967ArchMicrobiol(2016)198:53–69BourtzisK(2008)Wolbachia-basedtechnologiesforinsectpestpopulationcontrol.
AdvExpMedBiol627:104–113.
doi:10.
1007/978-0-387-78225-6_9BraytonKA,PalmerGH,LundgrenA,YiJ,BarbetAF(2002)Anti-genicvariationofAnaplasmamarginalemsp2occursbycombi-natorialgeneconversion.
MolMicrobiol43:1151–1159BrownlieJC,CassBN,RieglerM,WitsenburgJJ,Iturbe-OrmaetxeI,McGrawEA,O'NeillSL(2009)Evidenceformetabolicpro-visioningbyacommoninvertebrateendosymbiont,Wolbachiapipientis,duringperiodsofnutritionalstress.
PLoSPathog5:e1000368.
doi:10.
1371/journal.
ppat.
1000368BrutinelED,DeanAM,GralnickJA(2013)Descriptionofaribofla-vinbiosyntheticgenevariantprevalentinthephylumproteobac-teria.
JBacteriol195:5479–5486ChávezAS,FelsheimRF,KurttiTJ,KuPS,BraytonKA,MunderlohUG(2012)ExpressionpatternsofAnaplasmamarginaleMsp2variantschangeinresponsetogrowthincattle,andtickcellsversusmammaliancells.
PLoSOne7(4):e36012.
doi:10.
1371/journal.
pone.
0036012ComandatoreF,SasseraD,MontagnaM,KumarS,DarbyA,BlaxterMetal(2013)Phylogenomicsandanalysisofsharedgenessug-gestasingletransitiontomutualisminWolbachiaofnematodes.
GenomeBiolEvol5:1668–1674CordauxR,Michel-SalzatA,BouchonD(2001)Wolbachiainfectionincrustaceans:novelhostsandpotentialroutesforhorizontaltransmission.
JEvolBiol14:237–243CordauxR,PichonS,LingA,PerezP,DelaunayC,VavreF,BouchonD,GreveP(2008)Intensetranspositionalactivityofinsertionsequencesinanancientendosymbiont.
MolBiolEvol25:1889–1895.
doi:10.
1093/molbev/msn134CrossleyRA,GaskinDJ,HolmesK,MulhollandF,WellsJM,KellyDJ,vanVlietAH,WaltonNJ(2007)RiboflavinbiosynthesisisassociatedwithassimilatoryferricreductionandironacquisitionbyCampylobacterjejuni.
ApplEnvironMicrobiol73:7819–7825DedeineF,BandiC,BouletreauM,KramerL(2003)InsightsintoWolbachiaobligatorysymbiosis.
In:BourtzisK,MillerTA(eds)Insectsymbiosis.
CRCPress,Florida,pp267–282DellA,GaladariA,SastreF,HitchenP(2010)Similaritiesanddif-ferencesintheglycosylationmechanismsinprokaryotesandeukaryotes.
IntJMicrobiol.
doi:10.
1155/2010/148178FallonAM(2008)CytologicalpropertiesofanAedesalbopictusmos-quitocelllineinfectedwithWolbachiastrainwAlbB.
InVitroCellDevBiolAnim44:154–161.
doi:10.
1007/s11626-008-9090-4FallonAM,BaldridgeGD,HigginsLA,WitthuhnBA(2013)Wol-bachiafromtheplanthopperLaodelphaxstriatellusestablishesarobust,persistent,streptomycin-resistantinfectioninclonalmos-quitocells.
InVitroCellDevBiolAnim49:66–73.
doi:10.
1007/s11626-012-9571-3FallonAM,BaldridgeGD,CarrollEM,KurtzCM(2014)DepletionofhostcellriboflavinreducesWolbachiainculturedmosquitocells.
InVitroCellDevBiolAnim50:707–713.
doi:10.
1007/s11626-014-9758-xFelixC,PichonS,Braquart-VarnierC,BraigH,ChenL,GarrettRA,MartinG,GreveP(2008)CharacterizationandtranscriptionalanalysisoftwogeneclustersfortypeIVsecretionmachin-eryinWolbachiaofArmadillidiumvulgare.
ResMicrobiol159:481–485FloateKD,CoghlinPC,DosdallL(2011)AtestusingWolbachiabacteriatoidentifyEurasiansourcepopulationsofCabbageSeedPodWeevil,Ceutorhynchusobstrictus(Marsham),inNorthAmerica.
EnvironEntomol40:818–2011.
doi:10.
1603/EN10315GillAG,DarbyAC,MakepeaceBL(2014)Ironnecessity:thesecretofWolbachia'ssuccessPLoSNeglTropDis16:e3224.
doi:10.
1371/journal.
pntd.
0003224GillespieJJ,AmmermanNC,Dreher-LesnickSM,RahmanMS,Wor-leyMJ,SetubalJC,SobralBS,AzadAF(2009)AnanomaloustypeIVsecretionsysteminRickettsiaisevolutionarilycon-served.
PLoSOne4:e4833.
doi:10.
1371/journal.
pone.
0004833GillespieJJ,BraytonKA,WilliamsKP,QuevadoDiazMA,BrownWC,AzadAF,SobralBW(2010)PhylogenomicsrevealsadiverseRickettsialestypeIVsecretionsystem.
InfectImmun78:1809–1823HilgenboeckerK,HammersteinP,SchlattmannP,TelschowA,Wer-renJH(2008)HowmanyspeciesareinfectedwithWolbachiaAstatisticalanalysisofcurrentdata.
FEMSMicrobiolLett281:215–220HosokawaT,KogaR,KikuchiY,MengX-Y,FukatsuT(2010)Wol-bachiaasabacteriocyte-associatednutritionalmutualist.
ProcNatlAcadSciUSA107:769–774HotoppJC,LinM,MadupuR,CrabtreeSV,AngilouiSVetal(2006)Comparativegenomicsofemerginghumanehrlichiosisagents.
PLoSGenet2:e21JamesAC,DeanMD,McMahonME,BallardJW(2002)DynamicsofdoubleandsingleWolbachiainfectionsinDrosophilasimu-lansfromNewCaledonia.
Heredity88:182–189JinC,BarrientosA,TzagoloffA(2003)Yeastdihydroxybutanonephosphatesynthase,anenzymeoftheriboflavinbiosyntheticpathway,hasasecondunrelatedfunctioninexpressionofmito-chondrialrespiration.
JBiolChem278:14698–14703KambrisZ,CookPE,PhucHK,SinkinsSP(2009)Immuneactivationbylife-shorteningWolbachiaandreducedfilarialcompetenceinmosquitoes.
Science326:134–136.
doi:10.
1126/science.
1177531KlassonL,WestbergJ,SapountzisP,NaslundK,LutnaesY,DarbyAC,VenetiZ,ChenL,BraigHR,GarretRetal(2009)ThemosaicstructureoftheWolbachiawRistraininfectingDrosoph-ilasimulans.
ProcNatlAcadSciUSA106:5725–5730KnegtFH,MelloLV,ReisFC,SantosMT,VicentiniR,FerrazLF,OttoboniLM(2008)ribBandribBAgenesfromAcidithiobacil-lusferrooxidans:expressionlevelsunderdifferentgrowthcondi-tionsandphylogeneticanalysis.
ResMicrobiol159:423–431KoebnikR,LocherKP,VanGelderP(2000)Structureandfunctionofbacterialoutermembraneproteins:barrelsinanutshell.
MolMicrobiol37:239–253KremerN,VoroninD,CharifD,MavinguiP,MollereauB,VavreF(2009)Wolbachiainterfereswithferritinexpressionandironmetabolismininsects.
PLoSPathog5:e1000630.
doi:10.
1371/journal.
ppat.
1000630LiZ,CarlowCKS(2012)CharacterizationoftranscriptionfactorsthatregulatethetypeIVsecretionsystemandriboflavinbio-synthesisinWolbachiaofBrugiamalayi.
PLoSOne7:e51597.
doi:10.
1371/journal.
pone0051597LiuH,BaoW,LinM,NiuH,RikihisaY(2012)EhrlichiatypeIVsecretioneffectorECH0825istranslocatedtomitochondriaandcurbsROSandapoptosisbyupregulatingMnSOD.
CellMicro-biol14:1037–1050.
doi:10.
1111/j.
1462-5822.
2012.
01775.
xLoN,ParaskevopoulosC,BourtzisK,O'NeillSL,WerrenJH,Bor-densteinSR,BandiC(2007)Taxonomicstatusoftheintracel-lularbacteriumWolbachiapipientis.
IntJSystEvolMicrobiol57:654–657LockwoodS,VothDE,BraytonKA,BearePA,BrownWC,HeinzenRA,BroschatSL(2011)IdentificationofAnaplasmamarginaletypeIVsecretionsystemeffectorproteins.
PLoSOne6:e27724.
doi:10.
1371/journal.
pone.
0027724MasuiS,SasakiT,IshikawaH(2000)GenesforthetypeIVsecre-tionsysteminanintracellularsymbiont,Wolbachia,acausativeagentofvarioussexualalterationsinarthropods.
JBacteriol182:6529–6531McDonaghB,ReguejoR,Fuentes-AlmagroCA,OguetaS,BárcenaJA,PadillaCA(2011)Thiolredoxproteomicsidentifiesdiffer-entialtargetsofcytosolicandmitochondrialglutaredoxin-2iso-formsinSaccharomycescerevisiae.
ReversibleS-glutathionyla-tionofDHBPsynthase(RIB3).
JProteomics74:2487–249768ArchMicrobiol(2016)198:53–69MelnikowE,XuS,LiuJ,LiL,OksovY,GhedinEetal(2011)Inter-actionofaWolbachiaWSP-likeproteinwithanuclear-encodedproteinofBrugiamalayi.
IntJParasitol41:1053–1061MorrowJL,FrommerM,ShearmanDC,RieglerM(2014)Tropi-caltephritidfruitflycommunitywithhighincidenceofsharedWolbachiastrainsasplatformforhorizontaltransmis-sionofendosymbionts.
EnvironMicrobiol16:3622–3637.
doi:10.
1111/1462-2920.
12382NewtonLG,BordensteinSR(2011)Correlationsbetweenbacte-rialecologyandmobileDNA.
CurrMicrobiol62:198–208.
doi:10.
1007/s00284-010-9693-3NiuH,Kozjak-PavlovicV,RudelT,RikihisaY(2010)AnaplasmaphagocytophilumAts-1isimportedintohostcellmitochon-driaandinterfereswithapoptosisinduction.
PLoSPathog6:e1000774NodaH,KoizumiY,ZhangQ,DengK(2001a)InfectiondensityofWolbachiaandincompatibilitylevelintwoplanthopperspecies,LaodelphaxstriatellusandSogatellafurcifera.
InsectBiochemMolBiol31:727–737NodaH,MiyoshiT,ZhangQ,WatanabeK,DengK,HoshizakiS(2001b)Wolbachiainfectionsharedamongplanthoppers(Homoptera:Delphacidae)andtheirendoparasite(Strepsiptera:Elenchidae):aprobablecaseofinterspeciestransmission.
MolEcol10:2101–2106NodaH,MyoshiT,KoizumiY(2002)InvitrocultivationofWol-bachiaininsectandmammaliancelllines.
InVitroCellDevBiolAnim38:423–427NothaftH,SzymanskiCM(2010)Proteinglycosylationinbacteria:sweeterthanever.
NatRev8:775–778.
doi:10.
1038/nrmicro2383OfranY,RostB(2006)ISIS:interactionsitesidentifiedfromsequence.
Bioinformatics23:e13–e16.
doi:10.
1093/bioinformatics/bt303OhashiN,ZhiN,LinQ,RikihisaY(2002)CharacterizationandtranscriptionalanalysisofgeneclustersforatypeIVsecretionmachineryinhumangranulocyticandmonocyticehrlichiosisagents.
InfectImmun70:2128–2138O'NeillSL,PettigrewMM,SinkinsSP,BraigHR,AndreadisTG,TeshRB(1997)InvitrocultivationofWolbachiapipientisinanAedesalbopictuscellline.
InsectMolBiol6:33–39PanX,ZhouG,WuJ,BianG,LuP,RaikhelAS,XiZ(2012)Wol-bachiainducesreactiveoxygenspecies(ROS)-dependentacti-vationofthetollpathwaytocontroldenguevirusinthemos-quitoAedesaegypti.
ProcNatlAcadSciUSA109:E23–E31.
doi:10.
1073/pnas.
1116932108Perrot-MinnotMJ,GuoLR,WerrenJH(1996)SingleanddoubleinfectionswithWolbachiaintheparasiticwaspNasoniavitrip-ennis:effectsoncompatibility.
Genetics143:961–972PichonS,BouchonD,CordauxR,ChenL,GarretRA,GreveP(2009)ConservationofthetypeIVsecretionsystemthrough-outWolbachiaevolution.
BiochemBiophysResCommun385:557–562RancesE,VoroninD,Tran-VanV,MavanguiP(2008)GeneticandfunctionalcharacterizationofthetypeIVsecretionsysteminWolbachia.
JBacteriol190:5020–5030RaychoudhuryR,BaldoL,OliveiraDCSG,WerrenJH(2008)ModesofacquisitionofWolbachia:horizontaltransfer,hybridintro-gression,andconvergenceintheNasoniaspeciescomplex.
Evo-lution63:165–183.
doi:10.
1111/j.
1558-5646.
2008.
00533.
xRioR,HuY,AksoyS(2004)Strategiesofthehome-team:symbiosesexploitedforvector-bornediseasecontrol.
TrendsMicrobiol12:325–336.
doi:10.
1016/j.
tim.
2004.
05.
001SanogoYO,DobsonSL,BordensteinSR,NovakRJ(2007)Disrup-tionoftheWolbachiasurfaceproteingenewspBbyatranspos-ableelementinmosquitoesoftheCulexpipienscomplex(Dip-tera,Culicidae).
InsectMolBiol16:143–154SaridakiA,BourtzisK(2010)Wolbachia:morethanjustabugininsectgenitals.
CurrOpinMicrobiol13:67–72SarkarM,TroeseMJ,KearnsSA,YangT,ReneerDV,CarlyonJA(2008)AnaplasmaphagocytophilumMSP2(P44)-18pre-dominatesandismodifiedintomultipleisoformsinhumanmyeloidcells.
InfectImmun76:2090–2098.
doi:10.
1128/IAI.
01594-07SchulerH,BertheauC,EganSP,FederJL,RieglerM,Schlick-SteinerBC,SteinerFM,JohannesenJ,KernP,TubaK,Laka-tosF,KopplerK,ArthoferW,StaufferC(2013)EvidenceforarecenthorizontaltransmissionandspatialspreadofWolbachiafromendemicRhagoletiscerasi(Diptera:Tephritidae)toinva-siveRhagoletiscingulatainEurope.
MolEcol22:4101–4111.
doi:10.
1111/mec.
12362ShihKM,GerendayA,FallonAM(1998)CultureofmosquitocellsinEagle'smedium.
InVitroCellDevBiolAnim34:629–630SieversF,WilmA,DineenD,GibsonTJ,KarplusK,LiW,LopezR,McWilliamH,RemmertM,SodungJ,ThompsonJD,HigginsDG(2011)Fast,scalablegenerationofhigh-qualityproteinmul-tiplesequencealignmentsusingClustalOmega.
MolSystBiol7:539.
doi:10.
1038/msb.
2011.
75SinkinsSP,GouldF(2006)Genedrivesystemsforinsectdiseasevec-tors.
NatRevGenet7:427–435StrubingU,LuciusR,HoeraufA,PfarrKM(2010)Mitochondrialgenesforheme-dependentrespiratorychaincomplexesareup-regulatedafterdepletionofWolbachiafromfilarialnematodes.
IntJParasitol15:1193–1202.
doi:10.
1016/j.
ijpara.
2010.
03.
004SwoffordDL(2002)PAUP*.
Phylogeneticanalysisusingparsimony(*andothermethods).
Version4.
SinauerAssociates,SunderlandTaylorMJ,BandiC,HoeraufA(2005)Wolbachiabacterialendosym-biontsoffilarialnematodes.
AdvParasitol60:245–284UdayJ,PuttarajuHP(2012)ComparativeanalysisofWolbachiasur-faceproteininD.
melanoagster,A.
tabidaandB.
malayi.
Bioin-formation8(15):711–715.
doi:10.
6026/97320630008711WerrenJH,BaldoL,ClarkME(2008)Wolbachia:mastermanipula-torsofinvertebratebiology.
NatRevMicrobiol6:741–751WorstDJ,GerritsMM,Vandenbroucke-GraulsCM,KustersJG(1998)HelicobacterpyloriribBA-mediatedriboflavinproduc-tionisinvolvedinironacquisition.
JBacteriol180:1473–1479WuM,SunLV,VamathevanJ,RieglerM,DeboyR,BrownlieJC,McGrawE,MartinW,EsserCetal(2004)PhylogenomicsofthereproductiveparasiteWolbachiapipientiswMel:astream-linedgenomeoverrunbymobilegeneticelements.
PLoSBiol2:0327–0341WuB,NovelliJ,FosterJ,VaisvilaR,ConwayL,IngramJ,GanatraM,RaoAU,HamzaI,SlatkoB(2009)ThehemebiosyntheticpathwayoftheobligateWolbachiaendosymbiontofBrugiamalayiasapotentialanti-filarialdrugtarget.
PLoSNeglTropDis3:e475.
doi:10.
1371/journal.
pntd.
0000475YangC-H,XiaoJ-H,NiuL-M,MaG-C,CookJM,BianS-N,FuY-G,HuangD-H(2012)ChaosofWolbachiasequencesinsidethecompactfigsyconiaofFicusbenjamina(Ficus:Moraceae).
PLoSOne.
doi:10.
1371/journal.
pone.
0048882YurgelSN,RiceJ,DomreisE,LynchJ,SaN,QamarZ,RajamaniS,GaoM,RojeS,BauerWD(2014)Sinorhizobiummelilotiflavinsecretionandbacteria-hostinteraction:roleofthebifunc-tionalRibBAprotein.
MolPlant-MicrobeInteract27:437–445.
doi:10.
1094/MPMI-11-13-0338-RZabalouS,RieglerM,TheodorakopoulouM,StaufferC,SavakisC,BourtzisK(2004)Wolbachia-inducedcytoplasmicincompatibil-ityasameansforinsectpestpopulationcontrol.
ProcNatlAcadSciUSA101:15042–15045.
doi:10.
1073/pnas.
0403853101ZechnerEL,LangS,SchildbachJF(2012)Assemblyandmecha-nismsofbacterialtypeIVsecretionmachines.
PhilTransRSocB367:1073–1087.
doi:10.
1098/rstb.
2011.
020769ArchMicrobiol(2016)198:53–69ZhangKJ,HanX,HongXY(2013)VariousinfectionstatusandmolecularevidenceforhorizontaltransmissionandrecombinationofWolbachiaandCardiniumamongriceplanthoppersandrelatedspecies.
InsectSci3:329–344.
doi:10.
1111/j.
1744-7917.
2012.
01537.
xZugR,HammersteinP(2014)BadguysturnedniceAcriticalassessmentofWolbachiamutualismsinarthropodhosts.
BiolRev.
doi:10.
1111/brv.
12098ZugR,KoehnckeA,HammersteinP(2012)Epidemiologyinevo-lutionarytime:thecaseofWolbachiahorizontaltransmis-sionbetweenarthropodspecies.
JEvolBiol25:2149–2160.
doi:10.
1111/j.
1420-9101.
2012.
02601.
x
- transposasesodu.tw相关文档
- zidentyfikowanosodu.tw
- ostronocisodu.tw
- kszonysodu.tw
- monodispersesodu.tw
- sonecznychsodu.tw
- wybuchowesodu.tw
青果网络-618阿里云,腾讯云特惠优惠折上折!
官方网站:点击访问青果云官方网站活动方案:—————————–活动规则—————————1、选购活动产品并下单(先不要支付)2、联系我司在线客服修改价格或领取赠送时间3、确认价格已按活动政策修改正确后,支付订单,到此产品开设成功4、本活动产品可以升级,升级所需费用按产品原价计算若发生退款,按资源实际使用情况折算为产品原价再退还剩余余额! 美国洛杉矶CN2_GIACPU内存系统盘流量宽带i...
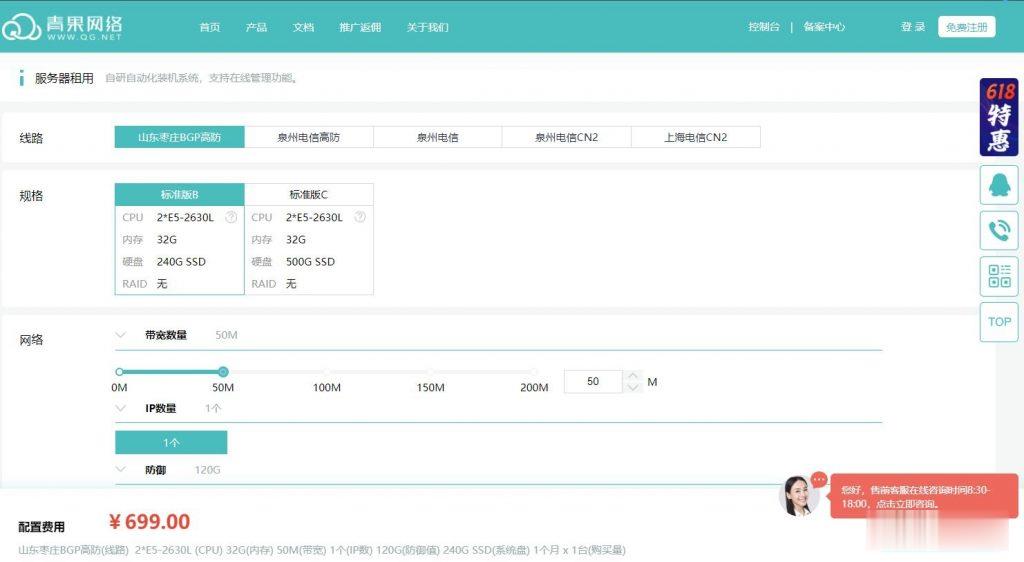
RAKsmartCloud服务器,可自定义配置月$7.59
RAKsmart商家一直以来在独立服务器、站群服务器和G口和10G口大端口流量服务器上下功夫比较大,但是在VPS主机业务上仅仅是顺带,尤其是我们看到大部分主流商家都做云服务器,而RAKsmart商家终于开始做云服务器,这次试探性的新增美国硅谷机房一个方案。月付7.59美元起,支持自定义配置,KVM虚拟化,美国硅谷机房,VPC网络/经典网络,大陆优化/精品网线路,支持Linux或者Windows操作...
RAKsmart含站群服务器/10G带宽不限流量首月半价
RAKsmart 商家估摸着前段时间服务器囤货较多,这两个月的促销活动好像有点针对独立服务器。前面才整理到七月份的服务器活动在有一些配置上比上个月折扣力度是大很多,而且今天看到再来部分的服务器首月半价,一般这样的促销有可能是商家库存充裕。比如近期有一些服务商挖矿服务器销售不好,也都会采用这些策略,就好比电脑硬件最近也有下降。不管如何,我们选择服务器或者VPS主机要本着符合自己需求,如果业务不需要,...
sodu.tw为你推荐
-
多家五星酒店回应网传名媛拼单在街上等公共场所拍到的视屏或者照片传到网上犯法吗?网易网盘关闭入口网易网盘怎么打不开了京沪高铁上市首秀京沪高铁怎么老是出问题?高铁的核心技术是中国自己的吗?小度商城小度在家智能屏Air性价比高吗?懂行的进~微信回应封杀钉钉微信大封杀什么时候结束www.hao360.cn搜狗360导航网址是什么lunwenjiancepaperfree论文检测怎样算合格xyq.163.cbg.comhttp://xyq.cbg.163.com/cgi-bin/equipquery.py?act=buy_show_equip_info&equip_id=475364&server_id=625 有金鱼贵吗?同ip域名什么是同主机域名同ip站点查询如何查看几个站是不是同IP