worldwidewww.13aaa.com
www.13aaa.com 时间:2021-04-08 阅读:()
SHORTREPORTOpenAccessFullsequenceanalysisandcharacterizationoftheSouthKoreanNorovirusGII-4variantCUK-3Jeong-WoongPark1,Sung-GeunLee2,Young-MinLee3,Weon-HwaJheong4,SangryeolRyu1*andSoon-YoungPaik2*AbstractBackground:Manyofresearchershavefocusedontheemergingpathogen,Norovirus,sinceitsfirstidentificationasthecausingagentofnonbacterialacutegastroenteritisinhumans.
Oneofthevirulencefactorsofnorovirus,thegreatgeneticdiversityattributedtopointmutationsandrecombinations,hasbroughtforththeresultofsignificantchangesinthecirculatingnorovirusgenotypepatterns.
Findings:Inrecognitionofthenecessityfortrackingandmonitoringofgeneticdiversity,anorovirusvariantamongthemostprevalentgenotypeGII-4,NorovirusHu/GII-4/CUK-3/2008/KR(CUK-3),wasisolatedfromstoolsamplesandanalyzedonthelevelofwholegenomesequence.
WholegenomesequenceanalysisrevealedthreeORFcompositesofthewholegenome,ORF1(5100bp),ORF2(1623bp),andORF3(807bp).
EachgeneticrelationshipofCUK-3variantanalysislocatedtheORF1(5,100bp)inClusterI,ORF2(1623bp)inClusterI(2006b),ORF3(807bp)inClusterI,andthewholegenomesequence(about5.
1kb)inClusterIinthephylogenetictree.
AndthephylogeneticanalysesshowedthesamelocationofCUK-3strainwiththeGII-4/2006bclusterinthephylogenetictree.
Conclusions:InThisstudy,afirstconcerningthefull-lengthsequenceofaNoVvariantinSouthKoreaismeaningfulinthatitcanbeusednotonlyasafull-lengthNoVvariantsequencestandardforfuturecomparisonstudies,butalsoasusefulmaterialforthepublichealthfieldbyenablingthediagnosis,vaccinedevelopment,andpredictionofnewemergingvariants.
Keywords:NorovirusSequence,PhylogenetictreeFindingsNoroviruses(NoVs)arethemostimportantvirusesthatcausenonbacterialacutegastroenteritisinhumans.
Inadditiontoincreasedsusceptibility,theelderlyareatincreasedriskformoreseverediseaseanddeath,asaretheveryyoungandtheimmunocompromised[1,2].
Theyaresmall,andnon-envelopedviruseswhichandbelongtothefamilyCaliciviridae,genusNorovirus.
Noroviruseshaveasinglepositive-strandNoVRNAgenomeofabout7.
6kbinsize.
Threeopenreadingframes(ORFs)havebeenidentifiedintheNoVgen-omes.
ORF1encodesapolyproteinthatiscleavedintosixnon-structural(NS)proteins,whichcarryaminoacidsequencemotifsconservedinNTPase,proteaseandRNA-dependentRNApolymerase(RdRp)[3,4].
ORF2encodesamajorstructuralprotein,ViralProtein(VP1),whichconsistoftwodomains-theshelldomain(S)andtheprotrudingarm(P)thatisagaindividedintotwosubdomains,P1andP2.
TheSdomainishighlycon-servedwhilethePdomainisvariable.
P2ofthePdomainishypervariableandcarriesimmuneandcellularrecognitionsites[5-7].
ORF3encodesminorcapsidpro-tein,VP2,whichisrichinbasicaminoacidsandispro-posedtohavearoleinviralstability[8,9]Recently,NoVswererecognizedasnovelemergentpathogens.
Themainrouteoftransmissionissuspectedtobeperson-to-person,butfoodandwater-borne*Correspondence:sangryu@snu.
ac.
kr;paik@catholic.
ac.
kr1DepartmentofFoodandAnimalBiotechnology,DepartmentofAgriculturalBiotechnology,CenterforAgriculturalBiomaterials,andResearchInstituteforAgricultureandLifeSciences,SeoulNationalUniversity,Seoul151-921,RepublicofKorea2DepartmentofMicrobiology,CollegeofMedicine,TheCatholicUniversityofKorea,Seoul137-701,RepublicofKoreaFulllistofauthorinformationisavailableattheendofthearticleParketal.
VirologyJournal2011,8:167http://www.
virologyj.
com/content/8/1/1672011Parketal;licenseeBioMedCentralLtd.
ThisisanOpenAccessarticledistributedunderthetermsoftheCreativeCommonsAttributionLicense(http://creativecommons.
org/licenses/by/2.
0),whichpermitsunrestricteduse,distribution,andreproductioninanymedium,providedtheoriginalworkisproperlycited.
transmissionisalsoimportant[1,10,11].
Accordingtonucleotidesequenceanalysisofthecapsidregions,noro-virusesareclassifiedintofivegenogroups,GItoGV,eachofwhichcanbefurtherdividedintoseveralclus-tersorgenotypes[12].
Amongthefivegenogroups,threegenogroups(GI,GIIandGIV)areknowntocauseclinicalillnessinhumans,andgenotypeGII-4hasbeenthepredominantcirculatingstraintothepresent[13].
Theerror-proneRNAreplicationandrecombinationbetweenvirusesiswhatdrivesnorovirusestoitsthegreatdiversity.
Furthermore,theaccumulatedmutationsofthehypervariableP2domainoftheVP1proteinpro-duceddifferentGII-4NoVs[14].
Themostrepresenta-tiveoftheresultingvariantGII-4strain,GII-4/2006b,with3nucleotideinsertionsintheP2domainatposi-tion6265,emergedinthesummerof2002whichleadtoamajorgastroenteritisoutbreakaswellasanepi-demicgastroenteritisworldwideinthewinterof2002/2003[15,16].
InSouthKorea,gastroenteritisoutbreaksbyGII-4/2006bvariantshavebeenreportedfromSep-temberof2007toJulyof2008hadbeenreported[17].
Inthispaper,thewholegenomesequenceofanotherisolatedvariantoftheemergingstraintype,theGII-4variant,wasanalyzedandcomparedwithothervariantstorevealthegeneticrelationshipandtopredicttheten-dencyofGII-4variantsinSouthKorea.
NoVpositive-stoolsamplewasisolatedfrompatientswithacutegastroenteritisinDaejeon,SouthKoreainNovember2008.
ThesamplewasobtainedfromtheWaterborneVirusBank(Seoul,Korea).
Thestoolsamplewasstoredat-70°.
TheviralgenomicRNAwasextractedfrom140μlof10%fecalsuspensionwiththeQIAampViralRNAMinikit(Qiagen,Hilden,Germany)inaccor-dancewiththemanufacturer'sinstructions.
ForthedetectionofNoV,reversetranscriptionPCR(RT-PCR)wasperformedwiththeOneStepRT-PCRkit(Qiagen,Hilden,Germany)withprimersbasedonthesequenceofthenoroviruscapsidregion(Table1).
Toanalyzethewholegenomesequenceofthedetectednorovirusstrain,reversetranscriptionPCR(RT-PCR)wasperformedwiththeOneStepRT-PCRkit(Qiagen,Hilden,Germany)with10pairsofnewlydesignedprimersets(Table1).
Tenfragmentswereamplified;eightfragmentsforORF1,onefragmentforORF2,andonefragmentforORF3.
ThePCRproductswerethenanalyzedbyelectrophoresison1.
5%agarosegelandethidiumbromidestaining.
Theamplifiedfrag-mentswerepurifiedwithHiYieldGel/PCRDNAExtractionkit(RBC,Taipei,Taiwan)fromthegel.
Then,theproductswereclonedintothepGEM-TEasyvector(Promega,USA)andweresequencedbyGenotech(Dae-jeon,SouthKorea).
SequencedataanalysisofthecompositesequenceoftenplasmidsalignedwithClustalWmethodusingtheDNAStarsoftware(DNAStarInc.
)revealedthatthewholegenomewascomposedofthreeORFs,5100bp(ORF1),1623bp(ORF2),and807bp(ORF3).
Thedendrogramswereconstructedwiththeneighbor-joiningmethod.
ThenucleotidesequencedatareportedinthisstudyhavebeendepositedintoGenBank(acces-sionnumber;FJ514242).
AcomparisonofthenucleotideandaminoacidsequencesoftheentireORF2withreferencestrainsshowedthatCUK-3strainhadthehighestsequencesimilaritytotheNoVGII-4referencestrainX76717.
ThenucleotidesimilaritywithCUK-3was60.
1-88.
0%withthereferencestrains.
Aproteindatabasesearch(BLASTX)fortheproductofORF2yielded59.
3-92.
0%similarity.
However,thecut-offvalueforaminoacidsimilaritybetweentheCUK-3variantandtheGII-4referencestrainsshowedlowidentity(92.
0%)forthegenomicsequences(Table2).
ToanalyzethegeneticrelationshipbetweentheCUK-3variantandtheothervariantsreportedworldwide,thesequencesofORFsequencesandthewholegenomewereprocessedwithmultiplesequencealignment.
EachanalysisofCUK-3locatedtheORF1(5,100bp)inClus-terI,ORF2(1623bp)inClusterI(2006b),andORF3(807bp)inClusterIaswellasthewholegenomesequence(about5.
1kb)inClusterI(Figure1).
AsshownonthecaseanalysisofORF1,ClusterIcon-tainsHiroshoma1,Kumamoto1,Saga1,Hokkaodo1,Aomori4,Miayagi5,Akita1,Aichi3andshowedthehighestsimilarityrange(98.
0~98.
9%)(Figure1A).
AsshownonthecaseanalysisofORF2,ClusterI(2006b)containsemergedtheGII-4varintsinJapanandshowedthehighestsimilarityrange(97.
7-99.
0%),whereasClusterII(2006a)showedlowsimilarityrange(91.
7-92.
3%)(Figure1B).
AsshownonthecaseanalysisofORF3,ClusterIshowedthehighestsimilarityrange(97.
9-99.
3%).
Inter-estingly,02/03groupcontainsemergedtheGII-4vaiantsinEuropeshowedmorehighersimilarityrange(93.
1-93.
6%)thanClusterIIcontainsemergedtheGII-4variantsinJapan(Figure1C).
Asshownonthecaseanalysisoffulllength,theCUK-3strainshowedthehighestsimilarity(98.
3%)withthe2006epidemicstrain,Aichi3/2006/JPstrain,whereasEhime/5-30/2005/JPstrainandLordsdale/1993/UKstrainshowedthelowestsimilarity(89.
2%)(Figure1D).
TheCUK-3straincanbeplacedonthesamebranchofthephylogenetictreeastheGII-4/2006bcluster,basedonthenucleotidesequenceoftheRdRpregionandtheaminoacidsequenceofthecapsidprotein(datanotshown).
Also,theanalysisoftheGDDmotif(AANNTG)oftheRdRpregionoftheGII-4variantsreportedworldwideindicatedthattheGDDmotifofCUK-3strainwassimilartothatoftheGII-4variantsofParketal.
VirologyJournal2011,8:167http://www.
virologyj.
com/content/8/1/167Page2of5theepidemicstrainsin2006.
Thesestrainswerecom-moninHongKong,Japan,andotherregionsofNorthEastAsiaduringthesameperiod[12,18].
Suchsimilari-tiesinanalysesbetweenCUK-3andGII-4/2006bstrainareevidenceofthereemergenceofGII-4variant.
InEuropeandAsia,mostnorovirusoutbreaksthatoccurredafter2002havebeencausedbyGII-4variantstrains(Figure2).
Inaddition,biennialemergencesofpandemicNoVGII-4variantshavebeenreportedin2002,2004,2006and2008[12,18,19].
TheseGII-4var-iantshavealsobeenidentifiedinSouthKoreaandarerapidlyspreadingworldwide.
AminoacidsequencesofthecapsidregionencodedbyORF2wascomparedwithatotalof39GII-4variantsreportedfromvariouscountriesincludingUS,UK,Japan,China,Germanyetc.
Dateof540aminoacidanalysesshowedthepatternofchangesoftwoaminoacidinfirstP1domain,aa238(SGII-4->2002TGII-4-2002FGII-4-2002PGII-4-2002PGII-4-1995YGII-4->2002FGII-4-2002EGII-4-2006TGII-4-1995QGII-4->2002)andaa504(PGII-4->2002QGII-4-<2002).
Theseanalyticdatashowedthattherehadalreadybeennumerousmutations,whichleadtotheproductionofawiderangeofvariants,alreadybefore2002andpro-ducedvariousvariants.
Andasshownonthecaseofaa394ofP2domainwhichinsertednovelaminoacid,GorS,andthensubstitutedtoTafter2006(datanotshown).
AlthoughpreviousstudieshaveindicatedthatthesequencefortheNoVcapsidregionisusefulforthediag-nosisofNoVvariants,nostudieshaveanalyzedtheRdRpregionoftheSouthKoreanNoVGII-4variant.
Thisisthefirstreportthatdescribesthefull-lengthsequenceofaNoVGII-4variantthatwasisolatedfromclinicalsamplesinSouthKorea.
Wesuggestthatthissequencecanbeusedasastandardforthecomparisonoffull-lengthNoVGII-4variantsequencewithotherstrains.
TheinformationacquiredfromwholegenomesequencinginthisstudycanbeusefulnotonlyforamoreaccuratediagnosesofNoVsbutalsoforthebasicresearchfortheelucidationgeneticfunctions.
Further-more,itwillbehelpfulforthepredictionofnewlyappearingpandemicvariantsthroughcomparisonwithGII-4variantsinneighboringcountries,fundamentalresearchforvaccinedevelopment,andeventually,forthefieldofpublichealththroughwiththeprovisionofnewemergingstrainsofNoV.
AcknowledgementsThisworkwassupportedbyMid-careerResearcherProgram(2008-0061575)andBasicScienceResearchProgram(2009-0062720)throughNRFgrantfundedbytheMEST.
Authordetails1DepartmentofFoodandAnimalBiotechnology,DepartmentofAgriculturalBiotechnology,CenterforAgriculturalBiomaterials,andResearchInstituteforAgricultureandLifeSciences,SeoulNationalUniversity,Seoul151-921,RepublicofKorea.
2DepartmentofMicrobiology,CollegeofMedicine,TheCatholicUniversityofKorea,Seoul137-701,RepublicofKorea.
3DepartmentofMicrobiology,CollegeofMedicineandMedicalResearchInstitute,ChungbukNationalUniversity,Cheongju,Chungcheongbuk-do361-763,RepublicofKorea.
4EnvironmentalInfrastructureResearchDepartment,NationalInstituteofEnvironmentalResearch,Incheon,404-708,RepublicofKorea.
Authors'contributionsSRRandSYPconceivedthisstudy.
YML,WHJanddesignedandconductedtheexperiments.
SGLandJWPanalyzedthesequencedataandcarriedoutthemolecularphylogeneticanalysis.
SGLandSYPwrotethemanuscript.
Allauthorsreadandapprovedthefinalmanuscript.
CompetinginterestsTheauthorsdeclarethattheyhavenocompetinginterests.
Received:28October2010Accepted:14April2011Published:14April2011ReferencesDonaldson,EF,LCLindesmith,ADLobue,andRSBaric.
2008.
Noroviruspathogenesis:mechanismsofpersistenceandimmuneevasioninhumanpopulations.
ImmunolRev225:190–211.
doi:10.
1111/j.
1600-065X.
2008.
00680.
x.
Yoon,JS,SGLee,SKHong,SALee,WHJheong,SSOh,MHOh,GPKo,CHLee,andSYPaik.
2008.
MolecularepidemiologyofnorovirusinfectionsinchildrenwithacutegastroenteritisinSouthKoreainNovember2005throughNovember2006.
JClinMicrobiol46:1474–1477.
doi:10.
1128/JCM.
02282-07.
Lee,YF,ANomoto,BMDetjen,andEWimmer.
1977.
AproteincovalentlylinkedtopoliovirusgenomeRNA.
ProcNatlAcadSciUSA74:59–63.
doi:10.
1073/pnas.
74.
1.
59.
Rueckert,RR,andEWimmer.
1984.
Systematicnomenclatureofpicornavirusproteins.
JVirol50:957–959.
Tan,M,PHuang,JMeller,WZhong,TFarkas,andXJiang.
2003.
MutationswithintheP2domainofnoroviruscapsidaffectbindingtohumanhisto-bloodgroupantigens:evidenceforabindingpocket.
JVirol77:12562–12571.
doi:10.
1128/JVI.
77.
23.
12562-12571.
2003.
Nilssson,MK,OHedlund,MThorhagen,GLarson,KJohansen,AEkspong,andLSvensson.
2003.
EvolutionofhumancalicivirusRNAinvivo:accumulationofmutationsintheprotrudingP2domainofthecapsidleadstostructuralchangesandpossiblyanewphenotype.
JVirol77:13117–13124.
doi:10.
1128/JVI.
77.
24.
13117-13124.
2003.
Lochridge,VP,KIJutila,JWGraff,andMEHardy.
2005.
EpitopesintheP2domainofnorovirusVP1recognizedbymonoclonalantibodiesthatblockcellinteractions.
JGenVirol86:2799–2806.
doi:10.
1099/vir.
0.
81134-0.
Glass,PJ,LJWhite,JMBull,ILeparc-Goffart,MEHardy,andMKEstes.
2000.
Norwalkvirusopenreadingframe3encodesaminorstructuralprotein.
JVirol74:6581–6591.
doi:10.
1128/JVI.
74.
14.
6581-6591.
2000.
Bertolotti-Ciariet,A,SECrawford,AMHutson,andMKEstes.
2003.
The3'endofNorwalkvirusmRNAcontainsdeterminantsthatregulatetheexpressionandstabilityoftheviralcapsidproteinVP1:anovelfunctionfortheVP2protein.
JVirol77:11603–11615.
doi:10.
1128/JVI.
77.
21.
11603-11615.
2003.
Kearney,K,JMenton,andJGMorgan.
2007.
Carlowvirus,a2002GII-4variantnorovirusstrainfromIreland.
VirolJ4:61.
doi:10.
1186/1743-422X-4-61.
LaRosa,G,MPourshaban,MIaconelli,andMMuscillo.
2008.
DetectionofgenogroupIVnorovirusesinenvironmentalandclinicalsamplesandpartialsequencingthroughrapidamplificationofcDNAends.
ArchVirol153:2077–2083.
doi:10.
1007/s00705-008-0241-4.
Motomura,K,TOka,MYokoyama,HNakamura,HMori,HOde,GSHansman,KKatayama,TKanda,TTanaka,NTakeda,andHSato.
2008.
Identificationofmonomorphicanddivergenthaplotypesinthe2006-2007norovirusGII/4epidemicpopulationbygenomewidetracingofevolutionaryhistory.
JVirol82:11247–11262.
doi:10.
1128/JVI.
00897-08.
Hale,A,KMattick,DLewis,MEstes,XJiang,JGreen,REglin,andDBrown.
2000.
DistinctepidemiologicalpatternsofNorwalk-likevirusinfection.
JMedVirol62:99–103.
doi:10.
1002/1096-9071(200009)62:13.
0.
CO;2-0.
Glass,RI,UDParashar,andMKEstes.
2009.
Norovirusgastroenteritis.
NEnglJMed361:1776–1785.
doi:10.
1056/NEJMra0804575.
Lopman,B,HVennema,EKohli,PPothier,ASanchez,ANegredo,JBuesa,ESchreier,MReacher,DBrown,JGray,MIturriza,CGallimore,BBottiger,KOHedlund,MTorvén,CHvonBonsdorff,LMaunula,MPoljsak-Prijatelj,JZimsek,GReuter,GSzücs,BMelegh,LSvennson,YvanDuijnhoven,andMKoopmans.
2004.
IncreaseinviralgastroenteritisoutbreaksinEuropeandepidemicspreadofnewnorovirusvariant.
Lancet363:682–688.
doi:10.
1016/S0140-6736(04)15641-9.
Allen,DJ,RNoad,DSamuel,JJGray,PRoy,andMIturriza-Gómara.
2009.
CharacterisationofaGII-4norovirusvariant-specificsurface-exposedsiteinvolvedinantibodybinding.
VirolJ6:150.
doi:10.
1186/1743-422X-6-150.
Chung,JY,THHan,SHPark,SWKim,andESHwang.
2010.
DetectionofGII-4/2006bVariantandRecombinantNorovirusesinChildrenWithAcuteGastroenteritis,SouthKorea.
JMedVirol82:146–152.
doi:10.
1002/jmv.
21650.
Ho,EC,PKCheng,AWLau,AHWong,andWWLim.
2007.
AtypicalnorovirusepidemicinHongKongduringsummerof2006causedbyanewgenogroupII/4variant.
JClinMicrobiol45:2205–2211.
doi:10.
1128/JCM.
02489-06.
Tu,ET,RABull,GEGreening,JHewitt,MJLyon,JAMarshall,CJMcIver,WDRawlinson,andPAWhite.
2008.
Epidemicsofgastroenteritisduring2006wereassociatedwiththespreadofnorovirusGII.
4variants2006aand2006b.
ClinInfectDis46:413–420.
doi:10.
1086/525259.
doi:10.
1186/1743-422X-8-167Citethisarticleas:Parketal.
:FullsequenceanalysisandcharacterizationoftheSouthKoreanNorovirusGII-4variantCUK-3.
VirologyJournal20118:167.
Parketal.
VirologyJournal2011,8:167http://www.
virologyj.
com/content/8/1/167Page5of5
Oneofthevirulencefactorsofnorovirus,thegreatgeneticdiversityattributedtopointmutationsandrecombinations,hasbroughtforththeresultofsignificantchangesinthecirculatingnorovirusgenotypepatterns.
Findings:Inrecognitionofthenecessityfortrackingandmonitoringofgeneticdiversity,anorovirusvariantamongthemostprevalentgenotypeGII-4,NorovirusHu/GII-4/CUK-3/2008/KR(CUK-3),wasisolatedfromstoolsamplesandanalyzedonthelevelofwholegenomesequence.
WholegenomesequenceanalysisrevealedthreeORFcompositesofthewholegenome,ORF1(5100bp),ORF2(1623bp),andORF3(807bp).
EachgeneticrelationshipofCUK-3variantanalysislocatedtheORF1(5,100bp)inClusterI,ORF2(1623bp)inClusterI(2006b),ORF3(807bp)inClusterI,andthewholegenomesequence(about5.
1kb)inClusterIinthephylogenetictree.
AndthephylogeneticanalysesshowedthesamelocationofCUK-3strainwiththeGII-4/2006bclusterinthephylogenetictree.
Conclusions:InThisstudy,afirstconcerningthefull-lengthsequenceofaNoVvariantinSouthKoreaismeaningfulinthatitcanbeusednotonlyasafull-lengthNoVvariantsequencestandardforfuturecomparisonstudies,butalsoasusefulmaterialforthepublichealthfieldbyenablingthediagnosis,vaccinedevelopment,andpredictionofnewemergingvariants.
Keywords:NorovirusSequence,PhylogenetictreeFindingsNoroviruses(NoVs)arethemostimportantvirusesthatcausenonbacterialacutegastroenteritisinhumans.
Inadditiontoincreasedsusceptibility,theelderlyareatincreasedriskformoreseverediseaseanddeath,asaretheveryyoungandtheimmunocompromised[1,2].
Theyaresmall,andnon-envelopedviruseswhichandbelongtothefamilyCaliciviridae,genusNorovirus.
Noroviruseshaveasinglepositive-strandNoVRNAgenomeofabout7.
6kbinsize.
Threeopenreadingframes(ORFs)havebeenidentifiedintheNoVgen-omes.
ORF1encodesapolyproteinthatiscleavedintosixnon-structural(NS)proteins,whichcarryaminoacidsequencemotifsconservedinNTPase,proteaseandRNA-dependentRNApolymerase(RdRp)[3,4].
ORF2encodesamajorstructuralprotein,ViralProtein(VP1),whichconsistoftwodomains-theshelldomain(S)andtheprotrudingarm(P)thatisagaindividedintotwosubdomains,P1andP2.
TheSdomainishighlycon-servedwhilethePdomainisvariable.
P2ofthePdomainishypervariableandcarriesimmuneandcellularrecognitionsites[5-7].
ORF3encodesminorcapsidpro-tein,VP2,whichisrichinbasicaminoacidsandispro-posedtohavearoleinviralstability[8,9]Recently,NoVswererecognizedasnovelemergentpathogens.
Themainrouteoftransmissionissuspectedtobeperson-to-person,butfoodandwater-borne*Correspondence:sangryu@snu.
ac.
kr;paik@catholic.
ac.
kr1DepartmentofFoodandAnimalBiotechnology,DepartmentofAgriculturalBiotechnology,CenterforAgriculturalBiomaterials,andResearchInstituteforAgricultureandLifeSciences,SeoulNationalUniversity,Seoul151-921,RepublicofKorea2DepartmentofMicrobiology,CollegeofMedicine,TheCatholicUniversityofKorea,Seoul137-701,RepublicofKoreaFulllistofauthorinformationisavailableattheendofthearticleParketal.
VirologyJournal2011,8:167http://www.
virologyj.
com/content/8/1/1672011Parketal;licenseeBioMedCentralLtd.
ThisisanOpenAccessarticledistributedunderthetermsoftheCreativeCommonsAttributionLicense(http://creativecommons.
org/licenses/by/2.
0),whichpermitsunrestricteduse,distribution,andreproductioninanymedium,providedtheoriginalworkisproperlycited.
transmissionisalsoimportant[1,10,11].
Accordingtonucleotidesequenceanalysisofthecapsidregions,noro-virusesareclassifiedintofivegenogroups,GItoGV,eachofwhichcanbefurtherdividedintoseveralclus-tersorgenotypes[12].
Amongthefivegenogroups,threegenogroups(GI,GIIandGIV)areknowntocauseclinicalillnessinhumans,andgenotypeGII-4hasbeenthepredominantcirculatingstraintothepresent[13].
Theerror-proneRNAreplicationandrecombinationbetweenvirusesiswhatdrivesnorovirusestoitsthegreatdiversity.
Furthermore,theaccumulatedmutationsofthehypervariableP2domainoftheVP1proteinpro-duceddifferentGII-4NoVs[14].
Themostrepresenta-tiveoftheresultingvariantGII-4strain,GII-4/2006b,with3nucleotideinsertionsintheP2domainatposi-tion6265,emergedinthesummerof2002whichleadtoamajorgastroenteritisoutbreakaswellasanepi-demicgastroenteritisworldwideinthewinterof2002/2003[15,16].
InSouthKorea,gastroenteritisoutbreaksbyGII-4/2006bvariantshavebeenreportedfromSep-temberof2007toJulyof2008hadbeenreported[17].
Inthispaper,thewholegenomesequenceofanotherisolatedvariantoftheemergingstraintype,theGII-4variant,wasanalyzedandcomparedwithothervariantstorevealthegeneticrelationshipandtopredicttheten-dencyofGII-4variantsinSouthKorea.
NoVpositive-stoolsamplewasisolatedfrompatientswithacutegastroenteritisinDaejeon,SouthKoreainNovember2008.
ThesamplewasobtainedfromtheWaterborneVirusBank(Seoul,Korea).
Thestoolsamplewasstoredat-70°.
TheviralgenomicRNAwasextractedfrom140μlof10%fecalsuspensionwiththeQIAampViralRNAMinikit(Qiagen,Hilden,Germany)inaccor-dancewiththemanufacturer'sinstructions.
ForthedetectionofNoV,reversetranscriptionPCR(RT-PCR)wasperformedwiththeOneStepRT-PCRkit(Qiagen,Hilden,Germany)withprimersbasedonthesequenceofthenoroviruscapsidregion(Table1).
Toanalyzethewholegenomesequenceofthedetectednorovirusstrain,reversetranscriptionPCR(RT-PCR)wasperformedwiththeOneStepRT-PCRkit(Qiagen,Hilden,Germany)with10pairsofnewlydesignedprimersets(Table1).
Tenfragmentswereamplified;eightfragmentsforORF1,onefragmentforORF2,andonefragmentforORF3.
ThePCRproductswerethenanalyzedbyelectrophoresison1.
5%agarosegelandethidiumbromidestaining.
Theamplifiedfrag-mentswerepurifiedwithHiYieldGel/PCRDNAExtractionkit(RBC,Taipei,Taiwan)fromthegel.
Then,theproductswereclonedintothepGEM-TEasyvector(Promega,USA)andweresequencedbyGenotech(Dae-jeon,SouthKorea).
SequencedataanalysisofthecompositesequenceoftenplasmidsalignedwithClustalWmethodusingtheDNAStarsoftware(DNAStarInc.
)revealedthatthewholegenomewascomposedofthreeORFs,5100bp(ORF1),1623bp(ORF2),and807bp(ORF3).
Thedendrogramswereconstructedwiththeneighbor-joiningmethod.
ThenucleotidesequencedatareportedinthisstudyhavebeendepositedintoGenBank(acces-sionnumber;FJ514242).
AcomparisonofthenucleotideandaminoacidsequencesoftheentireORF2withreferencestrainsshowedthatCUK-3strainhadthehighestsequencesimilaritytotheNoVGII-4referencestrainX76717.
ThenucleotidesimilaritywithCUK-3was60.
1-88.
0%withthereferencestrains.
Aproteindatabasesearch(BLASTX)fortheproductofORF2yielded59.
3-92.
0%similarity.
However,thecut-offvalueforaminoacidsimilaritybetweentheCUK-3variantandtheGII-4referencestrainsshowedlowidentity(92.
0%)forthegenomicsequences(Table2).
ToanalyzethegeneticrelationshipbetweentheCUK-3variantandtheothervariantsreportedworldwide,thesequencesofORFsequencesandthewholegenomewereprocessedwithmultiplesequencealignment.
EachanalysisofCUK-3locatedtheORF1(5,100bp)inClus-terI,ORF2(1623bp)inClusterI(2006b),andORF3(807bp)inClusterIaswellasthewholegenomesequence(about5.
1kb)inClusterI(Figure1).
AsshownonthecaseanalysisofORF1,ClusterIcon-tainsHiroshoma1,Kumamoto1,Saga1,Hokkaodo1,Aomori4,Miayagi5,Akita1,Aichi3andshowedthehighestsimilarityrange(98.
0~98.
9%)(Figure1A).
AsshownonthecaseanalysisofORF2,ClusterI(2006b)containsemergedtheGII-4varintsinJapanandshowedthehighestsimilarityrange(97.
7-99.
0%),whereasClusterII(2006a)showedlowsimilarityrange(91.
7-92.
3%)(Figure1B).
AsshownonthecaseanalysisofORF3,ClusterIshowedthehighestsimilarityrange(97.
9-99.
3%).
Inter-estingly,02/03groupcontainsemergedtheGII-4vaiantsinEuropeshowedmorehighersimilarityrange(93.
1-93.
6%)thanClusterIIcontainsemergedtheGII-4variantsinJapan(Figure1C).
Asshownonthecaseanalysisoffulllength,theCUK-3strainshowedthehighestsimilarity(98.
3%)withthe2006epidemicstrain,Aichi3/2006/JPstrain,whereasEhime/5-30/2005/JPstrainandLordsdale/1993/UKstrainshowedthelowestsimilarity(89.
2%)(Figure1D).
TheCUK-3straincanbeplacedonthesamebranchofthephylogenetictreeastheGII-4/2006bcluster,basedonthenucleotidesequenceoftheRdRpregionandtheaminoacidsequenceofthecapsidprotein(datanotshown).
Also,theanalysisoftheGDDmotif(AANNTG)oftheRdRpregionoftheGII-4variantsreportedworldwideindicatedthattheGDDmotifofCUK-3strainwassimilartothatoftheGII-4variantsofParketal.
VirologyJournal2011,8:167http://www.
virologyj.
com/content/8/1/167Page2of5theepidemicstrainsin2006.
Thesestrainswerecom-moninHongKong,Japan,andotherregionsofNorthEastAsiaduringthesameperiod[12,18].
Suchsimilari-tiesinanalysesbetweenCUK-3andGII-4/2006bstrainareevidenceofthereemergenceofGII-4variant.
InEuropeandAsia,mostnorovirusoutbreaksthatoccurredafter2002havebeencausedbyGII-4variantstrains(Figure2).
Inaddition,biennialemergencesofpandemicNoVGII-4variantshavebeenreportedin2002,2004,2006and2008[12,18,19].
TheseGII-4var-iantshavealsobeenidentifiedinSouthKoreaandarerapidlyspreadingworldwide.
AminoacidsequencesofthecapsidregionencodedbyORF2wascomparedwithatotalof39GII-4variantsreportedfromvariouscountriesincludingUS,UK,Japan,China,Germanyetc.
Dateof540aminoacidanalysesshowedthepatternofchangesoftwoaminoacidinfirstP1domain,aa238(SGII-4->2002TGII-4-2002FGII-4-2002PGII-4-2002PGII-4-1995YGII-4->2002FGII-4-2002EGII-4-2006TGII-4-1995QGII-4->2002)andaa504(PGII-4->2002QGII-4-<2002).
Theseanalyticdatashowedthattherehadalreadybeennumerousmutations,whichleadtotheproductionofawiderangeofvariants,alreadybefore2002andpro-ducedvariousvariants.
Andasshownonthecaseofaa394ofP2domainwhichinsertednovelaminoacid,GorS,andthensubstitutedtoTafter2006(datanotshown).
AlthoughpreviousstudieshaveindicatedthatthesequencefortheNoVcapsidregionisusefulforthediag-nosisofNoVvariants,nostudieshaveanalyzedtheRdRpregionoftheSouthKoreanNoVGII-4variant.
Thisisthefirstreportthatdescribesthefull-lengthsequenceofaNoVGII-4variantthatwasisolatedfromclinicalsamplesinSouthKorea.
Wesuggestthatthissequencecanbeusedasastandardforthecomparisonoffull-lengthNoVGII-4variantsequencewithotherstrains.
TheinformationacquiredfromwholegenomesequencinginthisstudycanbeusefulnotonlyforamoreaccuratediagnosesofNoVsbutalsoforthebasicresearchfortheelucidationgeneticfunctions.
Further-more,itwillbehelpfulforthepredictionofnewlyappearingpandemicvariantsthroughcomparisonwithGII-4variantsinneighboringcountries,fundamentalresearchforvaccinedevelopment,andeventually,forthefieldofpublichealththroughwiththeprovisionofnewemergingstrainsofNoV.
AcknowledgementsThisworkwassupportedbyMid-careerResearcherProgram(2008-0061575)andBasicScienceResearchProgram(2009-0062720)throughNRFgrantfundedbytheMEST.
Authordetails1DepartmentofFoodandAnimalBiotechnology,DepartmentofAgriculturalBiotechnology,CenterforAgriculturalBiomaterials,andResearchInstituteforAgricultureandLifeSciences,SeoulNationalUniversity,Seoul151-921,RepublicofKorea.
2DepartmentofMicrobiology,CollegeofMedicine,TheCatholicUniversityofKorea,Seoul137-701,RepublicofKorea.
3DepartmentofMicrobiology,CollegeofMedicineandMedicalResearchInstitute,ChungbukNationalUniversity,Cheongju,Chungcheongbuk-do361-763,RepublicofKorea.
4EnvironmentalInfrastructureResearchDepartment,NationalInstituteofEnvironmentalResearch,Incheon,404-708,RepublicofKorea.
Authors'contributionsSRRandSYPconceivedthisstudy.
YML,WHJanddesignedandconductedtheexperiments.
SGLandJWPanalyzedthesequencedataandcarriedoutthemolecularphylogeneticanalysis.
SGLandSYPwrotethemanuscript.
Allauthorsreadandapprovedthefinalmanuscript.
CompetinginterestsTheauthorsdeclarethattheyhavenocompetinginterests.
Received:28October2010Accepted:14April2011Published:14April2011ReferencesDonaldson,EF,LCLindesmith,ADLobue,andRSBaric.
2008.
Noroviruspathogenesis:mechanismsofpersistenceandimmuneevasioninhumanpopulations.
ImmunolRev225:190–211.
doi:10.
1111/j.
1600-065X.
2008.
00680.
x.
Yoon,JS,SGLee,SKHong,SALee,WHJheong,SSOh,MHOh,GPKo,CHLee,andSYPaik.
2008.
MolecularepidemiologyofnorovirusinfectionsinchildrenwithacutegastroenteritisinSouthKoreainNovember2005throughNovember2006.
JClinMicrobiol46:1474–1477.
doi:10.
1128/JCM.
02282-07.
Lee,YF,ANomoto,BMDetjen,andEWimmer.
1977.
AproteincovalentlylinkedtopoliovirusgenomeRNA.
ProcNatlAcadSciUSA74:59–63.
doi:10.
1073/pnas.
74.
1.
59.
Rueckert,RR,andEWimmer.
1984.
Systematicnomenclatureofpicornavirusproteins.
JVirol50:957–959.
Tan,M,PHuang,JMeller,WZhong,TFarkas,andXJiang.
2003.
MutationswithintheP2domainofnoroviruscapsidaffectbindingtohumanhisto-bloodgroupantigens:evidenceforabindingpocket.
JVirol77:12562–12571.
doi:10.
1128/JVI.
77.
23.
12562-12571.
2003.
Nilssson,MK,OHedlund,MThorhagen,GLarson,KJohansen,AEkspong,andLSvensson.
2003.
EvolutionofhumancalicivirusRNAinvivo:accumulationofmutationsintheprotrudingP2domainofthecapsidleadstostructuralchangesandpossiblyanewphenotype.
JVirol77:13117–13124.
doi:10.
1128/JVI.
77.
24.
13117-13124.
2003.
Lochridge,VP,KIJutila,JWGraff,andMEHardy.
2005.
EpitopesintheP2domainofnorovirusVP1recognizedbymonoclonalantibodiesthatblockcellinteractions.
JGenVirol86:2799–2806.
doi:10.
1099/vir.
0.
81134-0.
Glass,PJ,LJWhite,JMBull,ILeparc-Goffart,MEHardy,andMKEstes.
2000.
Norwalkvirusopenreadingframe3encodesaminorstructuralprotein.
JVirol74:6581–6591.
doi:10.
1128/JVI.
74.
14.
6581-6591.
2000.
Bertolotti-Ciariet,A,SECrawford,AMHutson,andMKEstes.
2003.
The3'endofNorwalkvirusmRNAcontainsdeterminantsthatregulatetheexpressionandstabilityoftheviralcapsidproteinVP1:anovelfunctionfortheVP2protein.
JVirol77:11603–11615.
doi:10.
1128/JVI.
77.
21.
11603-11615.
2003.
Kearney,K,JMenton,andJGMorgan.
2007.
Carlowvirus,a2002GII-4variantnorovirusstrainfromIreland.
VirolJ4:61.
doi:10.
1186/1743-422X-4-61.
LaRosa,G,MPourshaban,MIaconelli,andMMuscillo.
2008.
DetectionofgenogroupIVnorovirusesinenvironmentalandclinicalsamplesandpartialsequencingthroughrapidamplificationofcDNAends.
ArchVirol153:2077–2083.
doi:10.
1007/s00705-008-0241-4.
Motomura,K,TOka,MYokoyama,HNakamura,HMori,HOde,GSHansman,KKatayama,TKanda,TTanaka,NTakeda,andHSato.
2008.
Identificationofmonomorphicanddivergenthaplotypesinthe2006-2007norovirusGII/4epidemicpopulationbygenomewidetracingofevolutionaryhistory.
JVirol82:11247–11262.
doi:10.
1128/JVI.
00897-08.
Hale,A,KMattick,DLewis,MEstes,XJiang,JGreen,REglin,andDBrown.
2000.
DistinctepidemiologicalpatternsofNorwalk-likevirusinfection.
JMedVirol62:99–103.
doi:10.
1002/1096-9071(200009)62:13.
0.
CO;2-0.
Glass,RI,UDParashar,andMKEstes.
2009.
Norovirusgastroenteritis.
NEnglJMed361:1776–1785.
doi:10.
1056/NEJMra0804575.
Lopman,B,HVennema,EKohli,PPothier,ASanchez,ANegredo,JBuesa,ESchreier,MReacher,DBrown,JGray,MIturriza,CGallimore,BBottiger,KOHedlund,MTorvén,CHvonBonsdorff,LMaunula,MPoljsak-Prijatelj,JZimsek,GReuter,GSzücs,BMelegh,LSvennson,YvanDuijnhoven,andMKoopmans.
2004.
IncreaseinviralgastroenteritisoutbreaksinEuropeandepidemicspreadofnewnorovirusvariant.
Lancet363:682–688.
doi:10.
1016/S0140-6736(04)15641-9.
Allen,DJ,RNoad,DSamuel,JJGray,PRoy,andMIturriza-Gómara.
2009.
CharacterisationofaGII-4norovirusvariant-specificsurface-exposedsiteinvolvedinantibodybinding.
VirolJ6:150.
doi:10.
1186/1743-422X-6-150.
Chung,JY,THHan,SHPark,SWKim,andESHwang.
2010.
DetectionofGII-4/2006bVariantandRecombinantNorovirusesinChildrenWithAcuteGastroenteritis,SouthKorea.
JMedVirol82:146–152.
doi:10.
1002/jmv.
21650.
Ho,EC,PKCheng,AWLau,AHWong,andWWLim.
2007.
AtypicalnorovirusepidemicinHongKongduringsummerof2006causedbyanewgenogroupII/4variant.
JClinMicrobiol45:2205–2211.
doi:10.
1128/JCM.
02489-06.
Tu,ET,RABull,GEGreening,JHewitt,MJLyon,JAMarshall,CJMcIver,WDRawlinson,andPAWhite.
2008.
Epidemicsofgastroenteritisduring2006wereassociatedwiththespreadofnorovirusGII.
4variants2006aand2006b.
ClinInfectDis46:413–420.
doi:10.
1086/525259.
doi:10.
1186/1743-422X-8-167Citethisarticleas:Parketal.
:FullsequenceanalysisandcharacterizationoftheSouthKoreanNorovirusGII-4variantCUK-3.
VirologyJournal20118:167.
Parketal.
VirologyJournal2011,8:167http://www.
virologyj.
com/content/8/1/167Page5of5
- worldwidewww.13aaa.com相关文档
- stimuliwww.13aaa.com
- 2.45www.13aaa.com
- 负面www.13aaa.com
- wwwwww.13aaa.com
- disposedwww.13aaa.com
- Basedwww.13aaa.com
美国cera机房 2核4G 19.9元/月 宿主机 E5 2696v2x2 512G
美国特价云服务器 2核4G 19.9元杭州王小玉网络科技有限公司成立于2020是拥有IDC ISP资质的正规公司,这次推荐的美国云服务器也是商家主打产品,有点在于稳定 速度 数据安全。企业级数据安全保障,支持异地灾备,数据安全系数达到了100%安全级别,是国内唯一一家美国云服务器拥有这个安全级别的商家。E5 2696v2x2 2核 4G内存 20G系统盘 10G数据盘 20M带宽 100G流量 1...
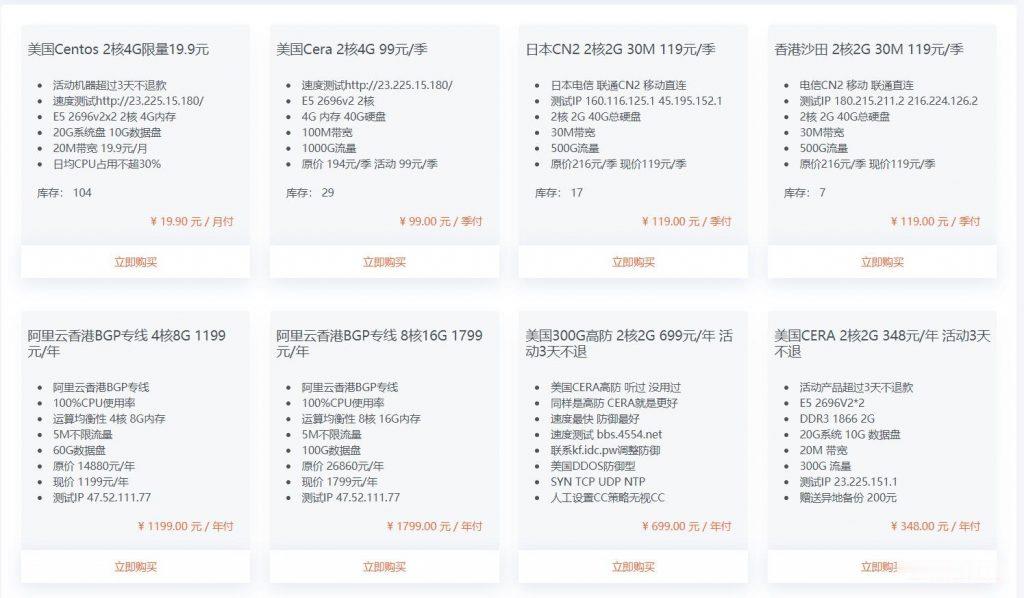
BuyVM商家4个机房的官方测试IP地址和测速文件
BuyVM 商家算是有一些年头,从早年提供低价便宜VPS主机深受广大网友抢购且也遭到吐槽的是因为审核账户太过于严格。毕竟我们国内的个人注册账户喜欢账户资料乱写,毕竟我们看英文信息有些还是比较难以识别的,于是就注册信息的时候随便打一些字符,这些是不能通过的。前几天,我们可以看到BUYVM商家有新增加迈阿密机房,而且商家有提供大硬盘且不限制流量的VPS主机,深受有一些网友的喜欢。目前,BUYVM商家有...
易探云香港云服务器价格多少钱1个月/1年?
易探云怎么样?易探云是目前国内少数优质的香港云服务器服务商家,目前推出多个香港机房的香港云服务器,有新界、九龙、沙田、葵湾等机房,还提供CN2、BGP及CN2三网直连香港云服务器。近年来,许多企业外贸出海会选择香港云服务器来部署自己的外贸网站,使得越来越多的用户会选择易探云作为网站服务提供平台。今天,云服务器网(yuntue.com)小编来谈谈易探云和易探云服务器怎么样?具体香港云服务器多少钱1个...
www.13aaa.com为你推荐
-
今日油条油条的由来及历史www.bbb336.comwww.zzfyx.com大家感觉这个网站咋样,给俺看看呀。多提意见哦。哈哈。www.haole012.comhttp://fj.qq.com/news/wm/wm012.htm 这个链接的视频的 第3分20秒开始的 背景音乐 是什么?125xx.comwww.free.com 是官方网站吗?www.544qq.COM跪求:天时达T092怎么下载QQip查询器怎么样查看自己电脑上的IP地址bbs2.99nets.com天堂1单机版到底怎么做javlibrary.comImage Library Sell Photos Digital Photos Photo Sharing Photo Restoration Digital Photos Photo Albums关键词分析怎么样分析关键词?官人放题求日本放题系列电影,要全集越多越好,求给力