methoxychrome
chrome18 时间:2021-01-31 阅读:()
SubjectIndexPagenumbersinitalicsrefertoformulaeandtablesacetaminophenols311o-acetanisidide311acetate,additiontoethyleneendgroupinpolyacetylenes201-incorporationintocycloheximide224f---eburiocoicacid146f.
---griseofulvin127ff.
---rugulosin101---streptidine378---variotin218,220acetate-malonatepathwaytopatulin85----polyacetylenes199acetate-propionaterouteinnystatinbio-synthesis228ff.
acetogenins88acetoxycycloheximide222acetyl-CoAinacetogeninbiosynthesis88--,precursorofpolyacetylenes199acetylenicbond,formationfromallenes198enolderivatives197f.
acetylene-dicarboxamide191-dicarboxylicacid,metabolisminEnterobacteriaceae196N-acetyl-glucosamine369--tetrahydrostreptobiosamine38815-N-acethyl-l5-N-hydroxyornithine,pre-cursorofferrichrome439acetyl-p-nitrophenylserinol32-phloroglucinol126C-acetylorsellinicacid89,95actidioneseecycloheximide222actinocin(2-amino-4,6-dimethyl-3-phen-oxazinone-1,9-dicarboxylicacid)227ff-,enzymaticsynthesis309ff.
ActinomycinA(I)282-B("antibioticX-45")276ff.
-C277ff,282-D(IV)277--chromophore(actinocin)277ff.
--inpolymyxinformation260---tyrothricineformation249actinomycinicacid315actinomycin-producingorganisms284,table285---,conditionsforcultivation284actinomycins,276ff.
,table278ff.
actinomycinsbiosynthesis,aminoacidincorporation,effectsofantibiotics(table)322precursors288,295--,inhibitionbyD-valine293ff.
--,relationtoproteinsynthesis320ff.
--,schemeofmechanism318f.
-,chemistry283ff.
-,chromophore,methylgroupsfrommethionine308-,controlledbiosynthesis286,293-,irrepressiblerepressorsofmRNAsyn-thesis331.
-,modeofaction283-,pentapeptides286,289-,-,formationdifferentfromproteinsynthesis320ff.
-,phenoxazinechromophore283ff.
-,roleinmetabolismofS.
antibioticus331.
-,therapeuticuses283-,"D-valine-D-alloisoleucine"-position283actiphenol222acumycin157,159,175adenine-D-allulosideseepsicofuranineadenine,incorporationintopsicofuramine403-,--toyocamycin407adenosine,conversiontocordycepin402,449S-adenosylmethionine115,167,305,392agrocybin191ff.
,192,194aklavinone102f.
,103albomycin17,439albomycetinseepicromycinalcohols,increasingoleandomycinpro-duction168aldgamycin157,159allenicbond,conversiontoacetylenicbond198D-alloisoleucine277,293,297f.
D-alloseintopsicofuranine403D-allose-6-phosphate449D-alluloseseeD-psicosealternariol89,127,128amaromycinseepicromycinN-amidino-2-deoxy-streptamine447SubjectIndex453N-amidino-scyllo-inosaminephosphate447N-amidino-streptamine447aminoacidanalogsintyrothricineforma-tion250--incorporationintoactinomycins288,303,320,322----penicillins3f.
aminoacids,aromatic,viridacinandbio-synthesisof106--inproductionofprodigiosin419,424aminoacyl-sRNA,attachmenttoriboso-mes,interferencebyedeine344D-or.
-aminoadipicacid1,3ff.
,5f.
,12or.
-aminoadipoylcysteine12d-(D-or.
-aminoadipoyl)-cysteinylvaline7,8,12f.
d-(L-or.
-aminoadipoyl)-sidechain5aminobenzoicacids311f.
7-aminocephalosporanicacid1,2,84-amino-dedimethylamino-7-chloran-hydrotetracycline1173'-amino-3'-deoxyadenosine4486-amino-9-(L-1,2-fucopyranoseenyl)-purineseeangustmycinA6-aminopenicillanicacid(6-APA)1,3f.
,8,124-aminopyrrolo-(2,3-d)-pyrimidine-p-D-ribofuranosideseetubercidinanacyclin194angolamycin157,159,175angustmycinA(6-amino-9-(L-1,2-fuco-pyranoseenyl)-purine)403f.
angustmycinCseepsicofuranineanhydro-7-chlortetracycline114anhydrooxytetracycline119,120anhydropenicillin10anisidines310ff.
anthranilicacid54,85,106anthraquinones,biosynthesis101ff.
,102antibioticformation,aminoacidsas"moderators"287antibioticPA-t05seeoleandomycin-PA-t08157,159,176-PA-133A157,159,176-PA-133B157,159,176-PA-148157,159,176-U-11,931(ethylanalogoflincomycin)354-U-11,973seeN-demethyl-lincomycin-U-20,943(S-ethylanalogoflinco-mycin)354f.
-U-21,699353f.
-X-45seeactinomycinB-ZN-6seefusidicacidD-apiose,formationfromD-glucose448L-arabinose361L-arcanose(2,6-dideoxy-3-o-methyl-3-C-methyl-L-xylohexose)154,155,158,168,178ff.
L-arginine361,379,381,447L-arginine-guanidino-uC447argomycinseepicromycinaromaticbiosynthesis,seealsoshikimicacidpathway--,startingfrompolyacetylenes208f.
arseniteinerythromycinbiosynthesis163f.
arylringinchloramphenicol38L-asparticacid361aspergillicacid29,43ff.
asperuloside137asterricacid96,97aucubin137aurantiogliocladin91f.
8-azaguanine,inhibitorofpolymyxinformation260azetidine-2-carboxylicacidinactino-mycinbiosynthesis291,300bacitracins,240ff.
,280-,biosynthesis,conditionsfor241ff.
-,-,inhibitors243f.
-,-,MnB+requirement241f.
-,-,relationtobasiccellularmechanisms245-,-byprotoplasts242-,effectsoncellmembranesofpro-ducingorganism244-,-othermicroorganisms244-,--sporulation245-asshuntmetabolite245benzophenonederivativesingriseofulvinfermentation127,129f.
benzylpenicillin(R)1,2,12biformyne-1(biformin)192,194,203biotin,ingriseofulvinbiosynthesis4402,2'-bipyrrolealdehyde4122,2'-bipyrroles449bisabolenes142bisdechlorogeodin130bluensomycin375bromogriseofulvin123,1242-butyl-3-ethylpyrrole413C1metabolism28calabarbean437caldariomycin98capillarine205capillene193carbomycinseemagnamycincarbondioxide38,41,226f.
carlinaoxide189,195,203f.
454SubjectIndexcarimbose(magnamycinlactone+mycaminose)170carotenoide134f.
catenulinseeparomycincellwalls,bacterial,mucopeptidesof,286cephalosporinC2ff.
,4,8--,chemicalformationfrompenicillin10-N(synnematinB)1-Pl146chalcomycin157ff.
,172f.
,176ff.
chalcose172chloramphenicol32ff.
-biosynthesis,media33--,feedbackcontrol34-,2,bromo-chloracetamidederivatives36-HC-incorporation37-,effectsonbiosynthesisofactino-mycin322ff.
bacitracin244edeine345f.
polymyxin260prodigiosin424tyrothricine249chlorinationoftetracyclines1173-chlorindole436,437chlorineactivationinchloramphenicolbiosynthesis417-chloro-4-aminodedimethylaminoanhydrotetracycline114,1177-chloro-6-demethyltetracycline113f.
3-chloro-4-(2'-nitro-3'-chloro-6'-hydroxy-phenyl)-pyrroleseeoxypyrrolnitrin3-chloro-4-{2'-nitro-3'-chlorophenyl)-pyrroleseepyrrolnitrin7-chlorotetracycline113ff.
-inedeineproduction345f.
--prodigiosinproduction424--tyrothricineformation249choline302,440chromogenicinduction416cicutoxin194cinnabarin318circulinA,B254,255-production,effectofmedia258cirramycins157,159,175citrinin98citronellal136citromycetin98,127,128-degradation99L-cladinose(2,6-dideoxY-3-0-methyl-3-C-methyl-L-ribohexose)154,155,158,162,167,178ff.
,441coccidioidomycosis,prodigiosinin411colimycin(neomycin)360coliphagefs343colistin(polymyxinE)255cordycepin(3'-deoxyadenosin)400f.
,402,448f.
-biosynthesis401f.
cordycepose(3-deoxyribose)401-,precursors402corynomycolicacid171crepenynicacid,polyacetyleneinter-mediate442cross-feedingins.
marcescens416cumulenes,intermediatesforthearomaticring444cyano-7-deazaadenineribonucleosideseetoyocamycincyclicethers,biogenesis442cyclitols384cycloheximide(actidione,naramycinA)222ff.
,222,223-,biosynthesis224f.
-degradation225f.
-dimethylcyclohexanonemoiety225f.
-glutarimidemoiety225f.
-,incorporationofprecursors225f.
cyclopaldicacidintropolonesynthesis90,93f.
cyclopenin106f.
cyclopenol,degradation106cyclopolicacid90,94cyclopropylcarbinylsysteminmarasmicacid141cysteinylvaline,cyclic7cytochromes,actinomycinsynthesis317-,productionincells24dammaranegroup148dammarenediols144,145darcanolide(lankanolide+lankavose)168daunomycin104daunomycinone(aglyconeofdauno-mycin)1047-deazapurinenucleosides,biosynthesisfrompurines405ff.
,scheme406dechlorogeodin97dechlorogriseofulvin123,124f.
decoyinine400,403f.
,448dehydro-7-chlorotetracycline114,119dehydrodechlorogriseofulvin131dehydrocycloheximide222dehydrogeodin97,130{-)-dehydrogriseofulvin124,127,129dehydromatricariaester201,204,441,443f.
--,conversiontothiophenes444dehydromatricarianol199"dehydrooxytetracycline"119SubjectIndex455sa,lla-dehydrotetracycline120D-demethyllincomycin(antibiotic1-11,973)354,3566-demethyltetracycline1133'-deoxyadenosinseecordycepindeoxyaspergillicacid486-deoxyerythronolideB440deoxystreptamine359,360,362-degradation365ff.
-,labellingpattern(table)365-,precursors368-,phosphateesters447deoxyviolacein77descarbamoylnovobiocin232,233,238O-desmethylnovobiocin232f,233desmycosin174d-desosamine(3,4,6-trideoxy-3-dime-thylamino-n-xylohexose)154,155,158,162,167,178ft.
dextromycinseeneomycindi-N-acetylneosaminol3672,4-diaminobutyricacid(DBA)255,262fIX,,B-diaminopimelicacid342dianthraquinones,biogenesisbypoly-acetate-route101f.
,(scheme)102diatetryne-3192-amide191ff.
,192,201-nitrile191ff.
,192,2012,3-dichloro-4-(2'-nitrophenyl)-pyrroleseeisopyrrolnitrin3,4-dihydro-6,8-dihydroxy-3,4,5-tri-methylisocoumarin-carboxylicacid,precursosofcitrinin98dihydronovobiocin237dihydrostreptomycin474L-dihydrostreptose(3-C-hydroxymethyl-5-deoxY-L-lyxofuranose)3742,3-dihydroxY-5,6-dimethyl-1,4-benzo-quinone91f.
3,5-dihydroxyphenylaceticacid1263,4-dihydroxyphenylalanine(DOPA)3172,4-dimethyl-3-acetylpyrrole425dimethylcyclohexanonemoietyofcycloheximide225f.
2,4-dimethyl-3-ethylpyrrole(krypto-pyrrole)414,425dimethylglycine3023,6-dimethyl-4-hydroXY-2-pyrone96,B,,B-dimethyllanthionine72,4-dimethylpyrrole414,4252,5-dimethylpyrrole4252,2'-dipyrrylmethanes449diterpenoids134f.
dithienyls443DNAsynthesis448dolichidial136drosophilinC192,194-D192,194eburicoicacid134,146f.
,147echinomycin286echinulin29edeine342ft.
,446-,bindingtoribosomes344-biosynthesisbycell-freesystem349---enzymefraction349ff.
inhibition350precursors350---intactcells344--andproteinsynthesis346ff.
---DNAsynthesisinvitro343-effectsonproteinsynthesisinvitro343-,mechanismofinvivoaction333-production,effectsofantibiotics345ff.
-,viricidalaction343emodin-5-methylester130endproductinhibitioninchloramphenicolsynthesis37enolphosphates,leadingtodiynes198enynicalcohols,furaneringfrom208erdin96erythromycin154ff.
,159,161,440f.
,176ft.
-,biogeneticscheme161-biosynthesis,alternatepathway164--,inhibitionbyarsenite163f.
-degradation165-effectonactinomycinbiosynthesis322,326-fermentation,conditions162-,glycosidationoferythronolides166-lactone(erythronolide),biosynthesis163,441--originof165-methylgroupfrommethionine392erythronolideseeerythromycinlactoneetamycin286,298ethionine117ethyl-IX-thiolincosamidine(ETL)354euphanegroup145,148euphol144,145falcarinone441trans-trans-farnesylpyrophosphate135,138f.
fattyacids,branchedchain,inbiosynthe-sisoferythromycin163ferrichrome17,439-,biosynthesis439ferrichromeA,fromt5-N-hydroxyorni-thine439ferrimycin17456SubjectIndexflagella,bacterial,e-N-methyl-lysinein301flavacol23,49flavipin90-degradation91flavomycinseeneomycinfolicanthine437formaldehyde,forD-psicose449formate18,386,440formylglycine18,20N-formyl-p-(methoxy)-styrylamine286-formyl-saIicylicacid85forosamine(2,3,4,6-tetrydeoxY-4-dime-thylamino-hexose154,155,158,173f.
,178ff.
fradiomycinseeneomycinframycetinseeneomycinD-fructose,incorporationintopsicofura-nine403D-fucoseinbiosynthesisofmacrolidesugars179fulvicacid125,127,128fungi,dermatophytic433-,pathogenetic449fumagillin152ff.
-biosyntheticscheme153fumigatin91ff.
fusarubin127,128fuscin99-synthesisfromacetate-polymalonateroute99f.
fusidicacid144ff.
-production145genipicacid137genipin137genipinicacid138gentamycins361gentisaldehyde85gentisicacid85,124gentisylalcohol85geodin96,118,130-biosynthesis97-hydrate97geodoxin97geranylgeranylpyrophosphate135geranylpyrophosphate135-unitinmycophenolicacid1oof.
gladiolicacid90gliotoxin29,439D-glucosamine362,365,368f.
,391D-glucose,formationofN-methyl-L-glu-cosamine389ff.
-,--streptose387ff.
-incorporation39(p-nitrophenylseri-noll,62(pyocyanin),171(magnamy-cins),361f.
,365,368(neomycins),391(streptomycin),401(cordycepose),403(psicofuranineanddecoyinine)L-glucose,notintostreptomycin391D-glucose-6-phosphate394D-glutamicacid361L-glutamicacid218ff.
(variotin),361(neomycin)glutarimideantibiotics222f.
-ringofcycloheximide225f.
glutathione7,286glutinone147,148glycerol57f.
glycine301-incorporation18(hadacidin),342(edeine),378(streptidine),421,424(prodigiosin)439(ferrichrome)glycolyticpathway395glyoxylicacid20gramicidin240f.
,245f.
,246,286-S245,246-effectonproteinsynthesis329f.
grifolin134,138f.
grisans123griseofulvin(2S,6'R)-7-chloro-4,6,2'-trimethoxy-6'-methylgris-2'en-3,4'-dione)96,123ff.
,440-biosyntheticpathway125ff.
(scheme),127---,halogenation131---,methylation130f.
-,degradation127-improvementofyieldbyN-methylcompounds126-production125griseomycins157,175L-gulose390hadacidin17ff.
,439-degradation18helvolicacid134,145f.
-production145f.
heptadecanoicacid163(erythromycin)hexa-N-acetylneomycin364L-histidine361(neomycin)L-homoarginine379(streptidine)homoorsellinicacid(2-ethyl-4,6-dihydro-benzoicacid)85humulene140hydronaphthacenicacidskeleton(tetra-cycline)115hydroxamates17,22f.
hydroxamicacids,biosynthesis4393-hydroxyanthranilicacid(3-HAA)309ff.
hydroxyaspergillicacid43p-hydroxybenzoicacid85-,precursorofAringofnovobiocin236.
SubjectIndex4574-hydroxy-2,2'bipyrrole-5-carboxalde-hyde(HBC)417p-hydroxycinnamicacid,precursorofBringofnovobiocin2357-hydroxycoumarinbiosynthesis23510-hydroxy-2-decenoicacid2925-hydroxy-Sa,11a-dehydrotetracycline120N-hydroxyglycine18,20ff.
,22D-IX-hydroxyisovalericacidinvalinomy-cin2693-hydroxykynurenineinactinomycin309ff.
hydroxylaminoacids(hadacidin)20ff.
2-hydroxy-3-methoxy-5,6-dimethyl-1,4-benzoquinone91f.
3-hydroxy-4-methylanthranilicacid(4-MHAA)inactinomycin3094-hydroxy-3-(3-methylbutyl)-benzoicacidindihydronovobiocin2374-hydroxy-6-methylpretetramid114ff.
hydroxymycinseeparomomycin2-hydroxyparidine311hydroxyphenazine586-N-hydroxyornithine23,439m-hydroxyphenylalanine(m-tyrosine)30hydroxyphthacenicacidskeletonintetracyclines,biosynthesisfromace-tate1153-hydroxyphthalicacid85hydroxY-L-proline286,298ff.
(actinomy-cin)2-hydroxypyridine-N-oxide43hydroxystreptomycin374,389L-hydroxystreptose(3-C-formyl-Iyxo-furanose)3745-hydroxytetracycline35,113,120illudinM140-S139f.
--,production139"Ilotycin"seeerythromycininactone222iridodial136iridomyrmecin136iron,inhibitionofhydroxymatesynthesis24isanicacid1893-isobutyl-6-(1-hydroxY-1-methyl-ethyl)-1-hydroxY-2(1H)pyrazinoneseemuta-aspergillicacid3-isobutyl-6-sec.
-butyl-2-hydroxy-pyrazine-1-oxideseeaspergillicacidisocycloheximide222,223isoiridomyrmecin136isonovobiocin232f.
,233isopenicillinN1,12isopentenylpyrophosphate134,135isophenoxazinesynthetase318isopyrrolnitrin(2,3-dichloro-4-(2'-nitro-phenyl)-pyrrole)434,435f.
isorhodomycinones102,103isotopecompetitiontechnique61isotyrosineinedeine3424-0-isovaleryl-mycaroseinleucomycins175junipal202kanamycinB360f.
5-keto-2,6-diamino-2,6-dideoxy-D-glu-cose,precursorofdiaminohexosefragmentinneomycins361ff.
4-ketodedimethylaminoanhydro-7-chlor-tetracycline114,1184-ketodedimethylaminoanhydrotetra-cycline114,117kojicacid440lachnophyllumester,189,B-lactam-dihydrothiazineringsystem(cephalosporins)12,B-lactam-thiazolidine(6-APA)ringsystem(penicillins)3,10,12lactate39lankanolide(aglyconeoflankamycin)168-biosynthesis169lankamycin156ff.
,168f.
,172,176ff.
D-lankavose(4,6-dideoxy-3-0-methyl-D-xylohexose)154,166,158,168,172,178ff.
lanosterol144,145(fusidicacid),147(eburicoicacid)D-leucine,46(aspergillicacid),261(poly-myxin)-,inhibitorofpolymyxinformation260L-leucine57(pyocyanin),172(L-myca-rose),261(polymyxin)cyclo-leucyl-leucyl23,49leukomycins157ff.
,175limonene136f.
lincocin(R)seelincomycinhydrochloridelincomycin363ff.
-biosynthesis,precursors355--,scheme357f.
-degradation356-hydrazinolysis353f.
-originofmethylgroups356macrolideantibiotics164ff.
--,biosynthesisbyacetate-polypro-pionatepolycondensations103--,-andnystatin228--,-,sugarparticipants158458SubjectIndexmacrolideantibiotics,biosynthesis,summary176f.
--,physicalconstants159--,producers(table)156f.
macrolidesugars155--,tentativepathwayforbiosynthe-sis178ff.
(flowsheet),178macrolides,6-deoxyhexosesattachedto154f.
,155-,oxidationto3-hydroxY-2.
4,6-tri-methylpimelicacidlactone169-,summaryofbuildingunits177magnamycin154ff.
,170,176ff.
-biosynthesisfromglucoseandsucci-nate171-,hydrolysis170-,precursors163malonate101(rugulosin).
127ff.
,440(griseofulvin),224f.
,(cycloheximide),(scheme)226;227(streptimidone)malonyl-CoA89(alternariol),115(tetra-cyclines),199(polyacetylenes)malonyl-pantotheine89(alternariol)maltol(3-hydroxy-2-methyl-4-pyrone),fromstreptose387maltose361(neomycins)D-mannose392(mannosidostreptomycin)mannosidostreptomycin(streptomycinB)375-,synthesis392f.
cx-mannosidostreptomycinase393marasin193-,enzymaticformationofalIenicsystem199marasmicacid140,141"matatabilactone"136matricariaester189,441"matromycin"seeoleandomycinmellein89"metatetrane"118f.
methionine,methyldonorinactinomycin308(table)cycloheximide224f.
fumagillin152lincomycin356N-methyl-L-glucosamine392f.
pyocyanine63streptose386variotin218,220-,precursorofpenicillinN63-methoxy-2,2'-bipyrrole-5-carboxalde-hyde4134-methoxy-2,2'-bipyrrole-5-carbox-aldehyde(MBC),precursorofprodi-giosin412ff.
,450N-methylalanine286N-methylalaninebiosynthesis301(actinomycin)N-methylaminoacidsinantibiotics3012-methyl-3-amylpyrrole(MAP),precur-sorofprodigiosin412ff.
2-methylanthraquinone1-methyl-4-ethyl-L-proline(ethylhygricacid),biosynthesis356N-methyl-L-glucosamine(N-methyl-2-amino-2-desoxy-L-glucopyranose)373f.
,378-,biosynthesis389ff.
-,mechanismofformation390-,penta-O-acetylderivative389methylgroups,migration444methylgroups,origininactinomycin308cycloheximide224f.
fumagillin152lincomycin356N-methyl-L-glucosamine392f.
myomycin329--,pyocyanine63streptose386U-21,699,356variotin218ff.
6-methylheptanoate(MHA)inpoly-myxins2552-methyl-3-heptyl-4-propylpyrrole4134-methyl-3-hydroxyanthranilicacid(4-MHAA)-peptidesinactinomycin310ff.
4-methyl-3-hydroxyanthraniloylpenta-peptideinactinomycinbiosynthesis292N-methyl-isoleucine292methylmalonate,incorporationintoerythromycins1662-methyl-5-methoxybenzoquinone91f.
N-methyl-mitomycinA676-(+)-methyloctanoate(MOA)inpoly-myxins255,262methylorsellinicacid92f.
6-methylpretetramid114ff.
3-methyl-DL-prolineinactinomycinbio-synthesis327ff.
4-methylproline3002-methylpyrrole413,425N-methyl-2.
4-quinazolinedionefromcyclopenol1066-methylsalicylicacid83,85,90,124,130--,acetylogs90--biosynthesis83f.
,88,90,92methyl-cx-thiolincosamidine(MTL)353ff.
methyltriaceticlactone95cx-methyl-DL-valine,inhibitorofpenicillinbiosynthesis296SubjectIndex459N-methyl-L-valineinactinomycin277,283,286f.
,296,301ff.
methymolide(aglyconofmethymycin)160methymycin154ff.
,160ff.
,176ff.
-biosynthesis,propionaterule160f.
mevalonate,incorporationintoeburicoicacid147-,--fumagillin152f.
-,--fuscine100-,--mycophenolicacid100-lactone,incorporationintofumagillin153miamycin157,159mitiromycin66mitomycinsA,B,C66ff.
-,biosynthesis68ff.
-,-,fermentationmedia69-,-,inhibitionbyDL-ethionine70-,-ofmitosanenucleus75-,-,reversalbymethionine71adenosylmethionine72-,effectonactinomycinbiosynthesis322,326-nucleus(mitosane)66mitosanemoietyofmitomycins66,75monoanthraquinones,biosynthesisbypolyacetateroute101.
(scheme),102monomycinseeparomycinmonoterpenoids134f.
cis-trans-muconicacid144mucomycinA143muta-aspergillicacid46D-mycaminose(3,6-diedeoxy-3-dimethyl-amino-D-glucose)154,155,158,170ff.
,174,178ff.
L-mycarose(2,6-dideoxy-3-C-methyl-L-ribohexose)154,155,162,170ff.
,174f.
,178ff.
,441-fromglucose1713-(X-L-mycarosylerythronolide440mycelianamide17,29,104,124f.
,134-degradation105-,formationofaromaticring105mycifradinseeneomycinmycinones103f.
D-mycinose(6-deoxY-2,3-di-O-methyl-D-allose)154,155,158,172ff.
,178ff.
mycobacillin271ff.
-,antifungalspectrum271-biosynthesisandmetabolism272,445--,effectofmedia272--,incorporationofaminoacids273446(table)--,inhibitionbychloramphenicol273--,noninterferenceofstreptomycindependence273mycobacillinbiosynthesis,roleofnucleo-tide-linkedpeptides273,446--,studieswithnon-producermutants273-,production272mycobactin17mycomycin193ff.
mycophenolicacid99--,derivationofsidechain100--,synthesisfromacetatepolymalo-nateroute99f.
mycosamineinnystatin228myo-inositolinstreptidine382ff.
,390---streptomycin447myo-inosose,inhibitionofstreptomycinproduction385myomycin392naramycinAseecycloheximide222-B222,223narbomycin156ff.
,169f.
,176ff.
-biogeneticrelationshiptoothermacrolides169neamine359ff.
nemotin192nemotinicacid192neoaspergillicacid23,48-production48neobiosamine359,365neohydroxyaspergillicacid48-production48neomethymycin155f.
,159,160,176ff.
neomycinA359ff.
-B359f.
-C359ff.
neomycins,biosynthesis,relationtocellwallsynthesis370-,degradationofsubunits(scheme)366-,effectonactinomycinbiosynthesis322,326-,precursors(table)363-,productionfromaminoacids361-,--sugars361-,subunits,biogeneticscheme362neosamineB359ff.
--labellingfromprecursors(table)365-C359,361f.
,364f.
,368,370--labellingfromprecursors(table)365,368neosamines,degradation365ff.
neospiramycins174neutramycin157,159,176niddamycin157,170f.
176ff.
o-nitroanisol311o-nitrobenzoicacid311nitrogroupinarylring,derivation40460SubjectIndexD-p-nitrophenylserinol(pNPS)inchlor-amphenicol35ff.
norprodigiosin413,415norvaline,inhibitoroftyrothricinforma-tion249noviose(Cringofnovobiocin)fromglu-cose231ff.
(scheme),233-biosynthesis,involvementofUDP-sugars232f.
vitaminB12232f.
novobiocicacid(B+Cringofnovobio-cin)231--,formation(scheme)236--,ringcoupling,enzymespecificity236novobiocin231f.
-biosynthesisofAring235f.
---Bring(coumarinmoiety)234f.
---Cringseenoviose--effectofprecursorsonrate235f.
--energyrequiringcouplingofringsystemsbycell-freeextracts236f.
--,proposedscheme237nystatin228ff.
-biosynthesisfromacetateandpropio-nate228ff.
-degradationtotiglicaldehyde228f.
-labellingpattern228f.
nystatinolide(aglyconofnystatin)228odyssicacid200oenanthotoxin194"Oleandocyn"seeoleandomycinoleandolide(aglyconeofoleandomycin)167f.
oleandomycin156ff.
,167ff.
,176ff.
-biogeneticscheme168-productionconditions168-triacetate(TAO)167L-oleandrose(2,6-dideoxY-2-0-methyl-L-arabohexose)154,155,158,167,178ff.
ommochrome(phenoxazinone)synthesis317oospolactone(C-methylderivativeofmellein)89orcinol(1,3-dihydroxY-4,5-dimethyl-benzene)91ff.
,138orcylaldehydeinstipitaticacid94DL-ornithineinprodigiosin421orsellinicacid82f.
,85,88,90,93,125,126,1384-oxo-6-deoxy-hexose,keyintermediateoferythromycinsugars1674-oxo-L-proline286-biosynthesis(actinomycin)298ff.
--distributionoflabel300oxygeninhydroxylaminogroup22oxypyrrolnitrin(3-chloro-4-(2'-nitro-3'-chloro-6'-hydroxy-phenyl)-pyrrole)434oxytetracyclinesee5-hydroxytetracyclinepalitanin128paromycins360,361patulin85,86,88,90,124D-penicillamine(D-p-thiovaline)4,296-,incorporationofvalines296penicillicacid85ff.
,87,88ff.
penicillins1ff.
-,aminoacidprecursors3ff.
-,chemicaltransformationtocephalo-sporins10-,6-APA-ringprecursors3f.
-,intermediates7f.
-,sidechainprecursors3f.
penicillinN1f.
,3ff.
-configurationofpenicillicacid296-effectonactinomycinbiosynthesis322,326pentadecanoicacidinerythromycins1632-pentenylpenicillinR1pentoseshuntinstreptosesynthesis387,395phenoxazinesynthesis,oxygenuptake316phenoxazinonesbyoxidativecondensa-tionfromo-aminophenols3174-MHAA-peptides315phenoxazinonesynthetase31Off.
--,inhibitionbyaminobenzenederi-vatives312ff.
chloramphenicol(table)3263-hydroxyanthranilicacidderivatives311ff.
--,metaleffects315f.
--,substratespecificity310phenoxy-methylphenylpenicillinmethylestersulphoxide10phenylalanine,precursorofchlorampheni-col35f.
-,--gliotoxin30-,--tropolones93-,--viridicatin106phenyl-et-mannosidehydrolysis393phenylpyrrolederivatives433ff.
---,halogenated434f.
phosphatidylethanolamine302phosphoenolpyruvate39o-phthaldialdehydes90f.
physostigmine437phytoene135picromycin154ff.
,155,158,176ff.
-biosynthesisseemethymycine159SubjectIndex461CSa-pigment(prodigiosin)413piperidinecarboxylicacidinactinomycinbiosynthesis288f.
,290f.
polyacetylenes189f.
,441f.
-,biogeneticcapabilities208f.
,209-,biosynthesis,feedingexperiments206ff.
-,cyclizationreactions203ff.
-,epoxide203f.
-,formationbydehydrogenation196-,furane203ff.
-,introductionoffunctionalgroups200.
-,mechanismoftriplebondformation195f.
-,originofC-skeleton199f.
-,spiroketalsof205,443-,thioether203ff.
poly-,B-ketides82ff.
,98,B-polyketomethylenechainfromacetate126ff.
--ingriseofulvinsynthesis127ff.
polymyxins254,255-,biosynthesis321-,-,inhibitors260ff.
-,-,mechanism260.
-,-andproteinsynthesis260-,--sporulation263f.
-,compositionof(table)254-,producingstrains255,(table)256-,production,media256f.
,258f.
-,-,carbonsource(table)256-,-,and2,4-diaminobutyricacid263-,-,nitrogensource(table)257-,-,andpH259-,-,andgrowthpolyphenoloxidase317porfiromycin66pretetramid116f.
pristimerin134,147f.
prodigiosin410ff.
,412,449f.
-biosynthesis414ff.
,(scheme)415--,aminoacidrequirement418--,isotopestudies420--,andthiamine420-,biosyntheticanalogs413f.
--,condensationreaction425--,environmentalfactors421-,chemistry411ff.
-,functions426-,fungicidalaction411-production,media418--,effectofair421f.
metals419f.
phosphate420temperature421f.
--,inhibitionbyantibiotics424-,sclerosingeffectinveins411prodigiosinsecondarymetaboliteinagedcultures426-,syntheticanalogs413-,water-solubleforms414-,cultureconditions450prodigiosin-likepigments,producedbyactinomycetes413L-prolineintoactinomycin277,291,298f.
,300-intoprodigiosin421ff.
propiolicacid189,195propionicacidintomacrolides160ff.
,163propylhygricacid(PHA)(trans-1-methyl-4-propyl-L-proline)353ff.
propylsuccinicacidfromlincomycin356proteinsynthesisandactinomycinforma-tion320ff.
---edeineformation346ff.
protetrone116protomycin222protozoa,inhibitionbyprodigiosin410psicofuranine(angustmycinC,adenine-alluloside)400,403ff.
,448f.
-biosynthesis403,449n-psicose(n-allulose)403-fromhexoses403-phosphate(n-allulose-6-phosphate)449puberulicacid93f.
puberulonicacid93f.
pulcherrimin17pulcherriminicacid23,50puromycin,effectonactinomycinbio-synthesis322,326-,--bacitracinbiosynthesis244-,--edeineproduction345f.
-,--tyrothricineformation249pyocyanine52-,carbonsources57,62-,degradation59-,productionbycellsuspensions56-,--growingcultures52-,-,media55pyrogallol85pyrrole425pyrrolnitrin(3-chloro-4-(2'-nitro-3'-chlorophenyl)-pyrrole)(=antibioticA10338)433f.
-antimicrobialspectrum433-incorporationoflabelledcompounds436f.
,437(table)-isolation434-mediumforproduction435-pathway437-precursors435f.
,-tradename:PYRO-ACE462SubjectIndexpyrromycinones102f.
,103pyrryldipyrrylmethene411ff.
pyruvateinpyocycaninesynthesis58--griseofulvin440pyoluteorin434quadrifidins193quadrilineatin90quinicacidinpyocyanin62o-quinonimineinphenoxazinesynthesis317D-quinovoseinmacrolidesugars179ramycinseefusidicacidrelomycin157,174f.
,176ff.
-fromtylosinbyreduction175ribonuclease,exogenous445ribose359,365(inneomycin),402(incordycepose)-,degradation366ff.
-,precursors(table)365,369D-ribose-5-phosphate,forD-psicose449ribosomes,bindingofedeine344RNAtemplate445"Romicil"seeoleandomycinroridins143rugulosin101-,C-C-couplingbyradicalmechanism101-,formationfromacetateandmalonate101saccharopine6sangivamycin407sarcosine277,287-biosynthesis301ff.
scyllo-inosamine,phosphateesters447scyllo-inositol(streptidine)384ff.
,390serine18,63,386,439f.
sepedonin95sesquiterpenoids134f.
siderochrome17shikimicacid37,60--pathway29,38,85,93,107,124spermidine344spiramycins157ff.
,173f.
,176ff.
spirodienoneintermediateinnovobiocinbiosynthesis234f.
,235all-trans-squalene145,147f.
steroids134f.
stipitaticacid94ff.
streptamine(1,3-diamino-2,4,5,6-tetra-hydroxycyclohexane)382,447-phosphateesters447streptidine(1,3-diguanidino-2,4,5,6-tetrahydroxycyclohexane)373f.
,378streptidinebiosynthesis378ff.
,447--,reactionsequence384ff.
streptimidone222,227streptobiosamine373f,388f.
streptomycin373ff.
,447f.
-biosynthesis,C-backbonefromglucose(table)383,(table)387,(table)388frommyo-inositol(table)383--,andgeneralmetabolism394f.
--,pathwaysinS.
griseus393--,sequentialsteps393ff.
-effectonactinomycinsynthesis322,326---mycobacillinsynthesis273---prodigiosinsynthesis424-,productionmedia376-,roleinlifeofproducers395L-streptose(3-C-formyl-5-deoXY-L-lyxo-furanose)373f.
,386-fromglucose386ff.
,448-,degradation388-,mechanismofformation387ff.
streptothricinBIseeneomycin-BlIseeneomycinstreptovitacinA,BandC.
222succinateintomagnamycins171--prodigiosin421,424N-succinyl-D-valine295f.
sulochrin97,124,129,130synthrophicpigmentationinS.
mar-cescens416tatiricacid189telomycin298terpenoidantibiotics134ff.
terphenylquinone30terthienyls444tertiomycins176a-terthienyl202tetracyclines113-,biosyntheticpathway(scheme)114-,effectonactinomycinbiosynthesis322,326-,-bacitracinbiosynthesis244-,--prodigiosinbiosynthesis424-,hydronaphthacenicacidskeleton115thioethers,polyacetylenic201.
,443f.
thiophenes,inpolyacetylenesbio-synthesis443f.
L-threonine277,283,293thymidinediphosphate-glucose(TDP-glucose)388,393--mannose(TDP-mannose)393--rhamnose(TDP-rhamnose)388SubjectIndex463tiglicaldehydefromnystatin228tirucal1anegroupofterpenoidanti-biotics145,148tirucallol144,145toyocamycin(cyano-7-deazaadenine-ribonucleoside)400,405,407-,adenineasprecursos407transamidinasesinstreptomycinproduction380f.
,447tricarboxylicacidcycleinstrepto-mycinformation395trichodermin142f.
trichodermol142f.
trichothecin141ff.
-production1423,5,7-trioxooctanoicacid126tripyrrylmethene411triterpenoids134f.
tropolones93ff.
trypacidin96n-tryptophan435f.
L-tryptophanintomitomycin75--violaceine78tryptophanmetabolism433tubercidin(4-aminopyrrolo-(2,3d)-pyrimidine-fJ-n-ribofuranoside)400f.
,405f.
,448tuberculostearicacid171tylosin157.
,174f.
,176ff.
-,reductiontorelomycin175tyrocidine240f.
,245f.
-,effectonproteinsynthesis329f.
L-tyrosineintomitomycin75--novobiocin234f.
--tropolones93--xanthocillin27f.
tyrothricin(mixtureofgramicidinandtyrocidine)247f.
-biosynthesis,conditions247.
--,inhibition250--,mechanism249f.
--,andproteinsynthesis250ubiquinone91f.
uridinediphosphate-N-acetylglucos-amine(UDP-NAG)inneomycinbiosynthesis369f.
usticacid89n-valineintoactinomycin277,293ff.
,296--valinomycin269-,utilizationbyS.
antibioticus293ff.
L-valine,conversionton-valine(actinomycinbiosynthesis)296-,intovalinomycin269-,utilizationinactinomycinbio-synthesis(table)297)valinomycin268ff.
-precursors269vanomycin,effectonactinomycinbiosynthesis322,326variotin216ff.
,217-biogenesisfromacetate217,218,220--pyrrolidonemoiety219-degradation216ff.
,(flowsheet)217-mediaforproduction217verrucarin143ff.
-Af42f.
,143-B143-,biogeneticscheme144-production143verrucarol143,144violacein77ff.
,450-,antibioticproperties79-,metabolicfunction78-,physiologicalrole78-,radiationprotection78-,toxiceffects79-,tryptophanin78viridicatin(2,3-dihydroXY-4-phenyl-quinoline)30,105f.
-biosynthesis,hypotheticalmecha-nism106f.
viridicatol(3'-hydroxyviridicatin)106f.
volucrisporin30xanthocillin26ff.
-,degradation27-,N-formyltoisonitrilegroups28-,precursors27xanthommatin317xanthones127,129ximenynicacid195zygomycinAseeparomomycinIndexofOrganismsActinidiapolygamaMIQ.
136Actinomycesscabies32Aerobacteraerogenes404,449Agaricaceae190Alcaligenesmetalcaligenes32Alternariatenuis89Anaphalis442Anthemissp.
201.
,204,443-tinctoria208Araliaceae190ArtemisiaannuaL.
202,207-capillaris193-vulgaris442Aspergillussp.
29-candidus98-Ilavus(PRL)43ff.
-fumigatusFRES.
83,92,96,152--mut.
helvola145-melleus89-niger433-oryzae45f.
-sclerotiorum(strainNRRL415)48ff.
-terreusTHOM.
93,96,98,130Bacillusanthracis286-brevis245,(Vm4)342ff.
,446-colistinus255,257-lichenilormis240-prodigiosus,seeSerratiamarcescens-pyocyaneus(Pseudomonasaeruginosa)52ff.
-subtilis79,271,289,364,433-polymyxa255ff.
Bacteriumprodigiosum,seeSerratiamarcescensBasidiomycetes190,192BidensconnatusMUHLENBG.
443BuphthalumsaliciloliumL.
444Blastomycesdermatidis216,449Caldariomyceslumago98Calycanthaceae437Calycanthusoccidentalis385,437Candidaalbicans433Carlinaacaulis204,208Celastrusdispermus(Maytenusdisper-mus)147-paniculatus147Celastrusscandens147-strigillosusNAKAI147CentaurearuthenicaLAM.
196,202,206Cephalosporiumsp.
1ff.
,146-lamellaecola144-mycophyllumIFO6615145ChaetomiumcochloidesPALL.
83f.
-globosum83ChamaemelumnobileL.
443ChimonanthusLINDL.
437ChromobacteriumprodigiosumseeSerratiamarcescens-violaceum78ff.
Chrysanthemummaximum200-segetumL.
207-serotinumL.
442f.
Cicutavirosa194Cladosporiumlulvum83Clitocybediatreta190,192-illudens,strain72027-5139f.
Clostridiumleseri32-welchii79Coccidioidesimmitis411Compositae190Coprinusquadrilidus190,193,200Cordycepsmilitaris401,448Coreopsissp.
203Cosmossulphureus200CousiniahystrixC.
A.
MAY200Curvularialunata83Daedaleajuniperinus202Dahliasp.
442Denhamiapitoosporoides147Drosophilasubatrata192Echinopssphaerocephalus208,444Entamoebahistolytica411Entoloma190Emericellopsissp.
1Escherichiacoli39,189,324,433Euphorbiaceae190Fomesol/icinalis146Fusidiumcoccineum144GenipaamericanaL.
137f.
Gibberellalujikuroi83IndexofOrganisms465Gliocladiumsp.
29-roseumBAINIER91Gnaphalium442Gramineae190Grifoliaconfluens138Helminthosporium448Hippocrateaindica147Histoplasmacapsulatum449Hydnum190lchthyothereterminalis194Iridomyrmexhumilis136Lactobacillusleichmanni402Lampteromycesjaponicus(KAWAM.
)SINGH139Lauraceae190Lentinusdactyloides146-degener91Leucopaxillusalbissimus191Loranthaceae190Marasmiuscongenus(Mo.
6890)140-ramealis193MicrococcusprodigiosusseeSerratiamarcescensMicrosporumaudouini216Minoliales190Mucorramannianus145Mycena190Mycobacteriumtuberculosis433MyrotheciumroridumS1135143-verrucaria143Neisseriameningitidis79Nitrobacteragilis21Nocardiaacidophila190-carallina175Oenanthecrocata193OidiodendronfuscumROBAK101Olacaceae190Opiliaceae190Oosporasp.
90-sulphurea-ochracea129Paecilomycespersicinus1-variotiBAINERvar.
antibioticus216f.
Papilionaceae190PastinacasativaL.
441Penicilliumsp.
29-albidumSoPP.
C.
M.
I.
40219124-aurantio-violaceum18ff.
--virensBIOURGE93Penicilliumbaarnense83f.
,103-brefeldianumDODGE124-brevicopactum83,90,101-brunneumUDAGAWA101-chrysogenumHf.
,433-citrinum98-cyclopiumWESTLING(NRRL1888)83,86,94,105-"estinogenum"97-frequentans99,128-gladioli100-griseofulvumDIERCKX83,104,124ff.
,440-islandicumSopp101-janczewskiiZAL.
[Ponigricans(BAI-NIER)THOM]124-janthinellum124-madriti83-meliniiTHIM.
124-notatumWESTLING26f.
-patulum(syn.
P.
urticae)BAIN83,85ff.
,124ff.
,440-paxilliivar.
echinulatum130-raciborskiiZAL.
124-raistrickiiSMITH124-rugulosumTHOM.
101-stipitatum83,93,95-urticae(=P.
patulum)125-viridicatumWESTLING105,107PhysostigmavenenosumBALF.
437Phytophthoracinnamoni191Polyporaceae190Polyporusanthracophilus146,199-biformis192-hisPidus146-sulphureus146-versicolor129Poriasp.
192-cocos146Pseudomonassp.
197-aeruginosa52f.
,434-aureojaciens434,436-bromoutilis434-methanica449-pyocyaneaseePs.
aeruginosa-pyrrociniasp.
n.
433Pristimeragrahami147-indica147Proteusvulgaris433Pycnoporussuccineus318Rhodotorulaglutinis191Rickettsiae32Saccharomycescerevisiae79Santalaceae190,195Santalumacuminatum199466IndexofOrganismsSerratiamarcescens410f.
,416,422,424,450--,mutant9-3-3412ff.
--,prodigiosin,nonproducermutants415ff.
--,sponteanouscolormutants415f.
--,temperature-inducedpigmenta-tion422,424Simarubaceae190Staphylococcusaureus289,324Streptomycessp.
1,102,154,224,269,279ff.
,316-albireticuli156,157,170,176-albus155,224-amakusaensis28-ambofaciens157,173-antibioticus156,157,167f.
,276ff.
-ardussp.
n.
66,67-aureofaciens115ff.
,157,281ff.
,285-aureoverticillatus450-bikiniensis157,172,447-bluensis374,447-caespitosus66f.
-chrysomallus277-cirratus157,175-djakartensis157,170-erythreus156,161ff.
,164ff.
,440-eurocidicus157,160,176.
-eurythermus157,175-felleus155ff.
-flaveolus285-flavochromogenes155f.
-flavus285-flavus-parvus285-fradiae157,174,281ff.
,359ff.
-fulvissimus268-griseocarnosus374-griseoflavus155f.
,157,175-griseolus157,175-griseus35,224,373ff.
,447f.
-halstedii156,164-hygroscopicus115,157,174,400,449--var.
angustmyceticus403f.
--var.
decoyicus403-lavendulae157,175-lincolnensisvar.
lincolnensis353ff.
-longisporusruber450-longissimus450-kitasatoensis157,175Streptomycesmacrosporeus159-melanochromogenes281ff.
-michiganensis285-naraensis224-narbonensis156,169-niveus231ff.
-noursei223,228-olivaceus156-parvulus279ff.
-pheochromogenes157--var.
chloromyceticus32-pulveraceus224-reticuli156.
,175--var.
protomycinus224-rimosusf.
paromomycinus224-rimosus35,115ff.
-ruber450-subrutilis374-tendae156-thermotolerans156,170-thioluteus21-tsusimaensis269-tubercidicus405-venezuelae32ff.
,155ff.
-verticillatus66,68ff.
-violaceoniger156,168Tagetessp.
194-erecta202-tenuifoliaCAV.
208Torulautilis433Trichodermaviride29f.
,143Tricholomagrammopodium199f.
Trichophytonasteroides433-interdigitale433-rubrum216,433Trichotheciumroseum141f.
TripterygiumregeliiSPRAGUEetTAKEDA147-wilfordiiHOOK147Trypanosomabrucei411-equiperdum411Umbelliferae190Ustilagosphaerogena439Valerianaceae190ViciafabaL.
191
---griseofulvin127ff.
---rugulosin101---streptidine378---variotin218,220acetate-malonatepathwaytopatulin85----polyacetylenes199acetate-propionaterouteinnystatinbio-synthesis228ff.
acetogenins88acetoxycycloheximide222acetyl-CoAinacetogeninbiosynthesis88--,precursorofpolyacetylenes199acetylenicbond,formationfromallenes198enolderivatives197f.
acetylene-dicarboxamide191-dicarboxylicacid,metabolisminEnterobacteriaceae196N-acetyl-glucosamine369--tetrahydrostreptobiosamine38815-N-acethyl-l5-N-hydroxyornithine,pre-cursorofferrichrome439acetyl-p-nitrophenylserinol32-phloroglucinol126C-acetylorsellinicacid89,95actidioneseecycloheximide222actinocin(2-amino-4,6-dimethyl-3-phen-oxazinone-1,9-dicarboxylicacid)227ff-,enzymaticsynthesis309ff.
ActinomycinA(I)282-B("antibioticX-45")276ff.
-C277ff,282-D(IV)277--chromophore(actinocin)277ff.
--inpolymyxinformation260---tyrothricineformation249actinomycinicacid315actinomycin-producingorganisms284,table285---,conditionsforcultivation284actinomycins,276ff.
,table278ff.
actinomycinsbiosynthesis,aminoacidincorporation,effectsofantibiotics(table)322precursors288,295--,inhibitionbyD-valine293ff.
--,relationtoproteinsynthesis320ff.
--,schemeofmechanism318f.
-,chemistry283ff.
-,chromophore,methylgroupsfrommethionine308-,controlledbiosynthesis286,293-,irrepressiblerepressorsofmRNAsyn-thesis331.
-,modeofaction283-,pentapeptides286,289-,-,formationdifferentfromproteinsynthesis320ff.
-,phenoxazinechromophore283ff.
-,roleinmetabolismofS.
antibioticus331.
-,therapeuticuses283-,"D-valine-D-alloisoleucine"-position283actiphenol222acumycin157,159,175adenine-D-allulosideseepsicofuranineadenine,incorporationintopsicofuramine403-,--toyocamycin407adenosine,conversiontocordycepin402,449S-adenosylmethionine115,167,305,392agrocybin191ff.
,192,194aklavinone102f.
,103albomycin17,439albomycetinseepicromycinalcohols,increasingoleandomycinpro-duction168aldgamycin157,159allenicbond,conversiontoacetylenicbond198D-alloisoleucine277,293,297f.
D-alloseintopsicofuranine403D-allose-6-phosphate449D-alluloseseeD-psicosealternariol89,127,128amaromycinseepicromycinN-amidino-2-deoxy-streptamine447SubjectIndex453N-amidino-scyllo-inosaminephosphate447N-amidino-streptamine447aminoacidanalogsintyrothricineforma-tion250--incorporationintoactinomycins288,303,320,322----penicillins3f.
aminoacids,aromatic,viridacinandbio-synthesisof106--inproductionofprodigiosin419,424aminoacyl-sRNA,attachmenttoriboso-mes,interferencebyedeine344D-or.
-aminoadipicacid1,3ff.
,5f.
,12or.
-aminoadipoylcysteine12d-(D-or.
-aminoadipoyl)-cysteinylvaline7,8,12f.
d-(L-or.
-aminoadipoyl)-sidechain5aminobenzoicacids311f.
7-aminocephalosporanicacid1,2,84-amino-dedimethylamino-7-chloran-hydrotetracycline1173'-amino-3'-deoxyadenosine4486-amino-9-(L-1,2-fucopyranoseenyl)-purineseeangustmycinA6-aminopenicillanicacid(6-APA)1,3f.
,8,124-aminopyrrolo-(2,3-d)-pyrimidine-p-D-ribofuranosideseetubercidinanacyclin194angolamycin157,159,175angustmycinA(6-amino-9-(L-1,2-fuco-pyranoseenyl)-purine)403f.
angustmycinCseepsicofuranineanhydro-7-chlortetracycline114anhydrooxytetracycline119,120anhydropenicillin10anisidines310ff.
anthranilicacid54,85,106anthraquinones,biosynthesis101ff.
,102antibioticformation,aminoacidsas"moderators"287antibioticPA-t05seeoleandomycin-PA-t08157,159,176-PA-133A157,159,176-PA-133B157,159,176-PA-148157,159,176-U-11,931(ethylanalogoflincomycin)354-U-11,973seeN-demethyl-lincomycin-U-20,943(S-ethylanalogoflinco-mycin)354f.
-U-21,699353f.
-X-45seeactinomycinB-ZN-6seefusidicacidD-apiose,formationfromD-glucose448L-arabinose361L-arcanose(2,6-dideoxy-3-o-methyl-3-C-methyl-L-xylohexose)154,155,158,168,178ff.
L-arginine361,379,381,447L-arginine-guanidino-uC447argomycinseepicromycinaromaticbiosynthesis,seealsoshikimicacidpathway--,startingfrompolyacetylenes208f.
arseniteinerythromycinbiosynthesis163f.
arylringinchloramphenicol38L-asparticacid361aspergillicacid29,43ff.
asperuloside137asterricacid96,97aucubin137aurantiogliocladin91f.
8-azaguanine,inhibitorofpolymyxinformation260azetidine-2-carboxylicacidinactino-mycinbiosynthesis291,300bacitracins,240ff.
,280-,biosynthesis,conditionsfor241ff.
-,-,inhibitors243f.
-,-,MnB+requirement241f.
-,-,relationtobasiccellularmechanisms245-,-byprotoplasts242-,effectsoncellmembranesofpro-ducingorganism244-,-othermicroorganisms244-,--sporulation245-asshuntmetabolite245benzophenonederivativesingriseofulvinfermentation127,129f.
benzylpenicillin(R)1,2,12biformyne-1(biformin)192,194,203biotin,ingriseofulvinbiosynthesis4402,2'-bipyrrolealdehyde4122,2'-bipyrroles449bisabolenes142bisdechlorogeodin130bluensomycin375bromogriseofulvin123,1242-butyl-3-ethylpyrrole413C1metabolism28calabarbean437caldariomycin98capillarine205capillene193carbomycinseemagnamycincarbondioxide38,41,226f.
carlinaoxide189,195,203f.
454SubjectIndexcarimbose(magnamycinlactone+mycaminose)170carotenoide134f.
catenulinseeparomycincellwalls,bacterial,mucopeptidesof,286cephalosporinC2ff.
,4,8--,chemicalformationfrompenicillin10-N(synnematinB)1-Pl146chalcomycin157ff.
,172f.
,176ff.
chalcose172chloramphenicol32ff.
-biosynthesis,media33--,feedbackcontrol34-,2,bromo-chloracetamidederivatives36-HC-incorporation37-,effectsonbiosynthesisofactino-mycin322ff.
bacitracin244edeine345f.
polymyxin260prodigiosin424tyrothricine249chlorinationoftetracyclines1173-chlorindole436,437chlorineactivationinchloramphenicolbiosynthesis417-chloro-4-aminodedimethylaminoanhydrotetracycline114,1177-chloro-6-demethyltetracycline113f.
3-chloro-4-(2'-nitro-3'-chloro-6'-hydroxy-phenyl)-pyrroleseeoxypyrrolnitrin3-chloro-4-{2'-nitro-3'-chlorophenyl)-pyrroleseepyrrolnitrin7-chlorotetracycline113ff.
-inedeineproduction345f.
--prodigiosinproduction424--tyrothricineformation249choline302,440chromogenicinduction416cicutoxin194cinnabarin318circulinA,B254,255-production,effectofmedia258cirramycins157,159,175citrinin98citronellal136citromycetin98,127,128-degradation99L-cladinose(2,6-dideoxY-3-0-methyl-3-C-methyl-L-ribohexose)154,155,158,162,167,178ff.
,441coccidioidomycosis,prodigiosinin411colimycin(neomycin)360coliphagefs343colistin(polymyxinE)255cordycepin(3'-deoxyadenosin)400f.
,402,448f.
-biosynthesis401f.
cordycepose(3-deoxyribose)401-,precursors402corynomycolicacid171crepenynicacid,polyacetyleneinter-mediate442cross-feedingins.
marcescens416cumulenes,intermediatesforthearomaticring444cyano-7-deazaadenineribonucleosideseetoyocamycincyclicethers,biogenesis442cyclitols384cycloheximide(actidione,naramycinA)222ff.
,222,223-,biosynthesis224f.
-degradation225f.
-dimethylcyclohexanonemoiety225f.
-glutarimidemoiety225f.
-,incorporationofprecursors225f.
cyclopaldicacidintropolonesynthesis90,93f.
cyclopenin106f.
cyclopenol,degradation106cyclopolicacid90,94cyclopropylcarbinylsysteminmarasmicacid141cysteinylvaline,cyclic7cytochromes,actinomycinsynthesis317-,productionincells24dammaranegroup148dammarenediols144,145darcanolide(lankanolide+lankavose)168daunomycin104daunomycinone(aglyconeofdauno-mycin)1047-deazapurinenucleosides,biosynthesisfrompurines405ff.
,scheme406dechlorogeodin97dechlorogriseofulvin123,124f.
decoyinine400,403f.
,448dehydro-7-chlorotetracycline114,119dehydrodechlorogriseofulvin131dehydrocycloheximide222dehydrogeodin97,130{-)-dehydrogriseofulvin124,127,129dehydromatricariaester201,204,441,443f.
--,conversiontothiophenes444dehydromatricarianol199"dehydrooxytetracycline"119SubjectIndex455sa,lla-dehydrotetracycline120D-demethyllincomycin(antibiotic1-11,973)354,3566-demethyltetracycline1133'-deoxyadenosinseecordycepindeoxyaspergillicacid486-deoxyerythronolideB440deoxystreptamine359,360,362-degradation365ff.
-,labellingpattern(table)365-,precursors368-,phosphateesters447deoxyviolacein77descarbamoylnovobiocin232,233,238O-desmethylnovobiocin232f,233desmycosin174d-desosamine(3,4,6-trideoxy-3-dime-thylamino-n-xylohexose)154,155,158,162,167,178ft.
dextromycinseeneomycindi-N-acetylneosaminol3672,4-diaminobutyricacid(DBA)255,262fIX,,B-diaminopimelicacid342dianthraquinones,biogenesisbypoly-acetate-route101f.
,(scheme)102diatetryne-3192-amide191ff.
,192,201-nitrile191ff.
,192,2012,3-dichloro-4-(2'-nitrophenyl)-pyrroleseeisopyrrolnitrin3,4-dihydro-6,8-dihydroxy-3,4,5-tri-methylisocoumarin-carboxylicacid,precursosofcitrinin98dihydronovobiocin237dihydrostreptomycin474L-dihydrostreptose(3-C-hydroxymethyl-5-deoxY-L-lyxofuranose)3742,3-dihydroxY-5,6-dimethyl-1,4-benzo-quinone91f.
3,5-dihydroxyphenylaceticacid1263,4-dihydroxyphenylalanine(DOPA)3172,4-dimethyl-3-acetylpyrrole425dimethylcyclohexanonemoietyofcycloheximide225f.
2,4-dimethyl-3-ethylpyrrole(krypto-pyrrole)414,425dimethylglycine3023,6-dimethyl-4-hydroXY-2-pyrone96,B,,B-dimethyllanthionine72,4-dimethylpyrrole414,4252,5-dimethylpyrrole4252,2'-dipyrrylmethanes449diterpenoids134f.
dithienyls443DNAsynthesis448dolichidial136drosophilinC192,194-D192,194eburicoicacid134,146f.
,147echinomycin286echinulin29edeine342ft.
,446-,bindingtoribosomes344-biosynthesisbycell-freesystem349---enzymefraction349ff.
inhibition350precursors350---intactcells344--andproteinsynthesis346ff.
---DNAsynthesisinvitro343-effectsonproteinsynthesisinvitro343-,mechanismofinvivoaction333-production,effectsofantibiotics345ff.
-,viricidalaction343emodin-5-methylester130endproductinhibitioninchloramphenicolsynthesis37enolphosphates,leadingtodiynes198enynicalcohols,furaneringfrom208erdin96erythromycin154ff.
,159,161,440f.
,176ft.
-,biogeneticscheme161-biosynthesis,alternatepathway164--,inhibitionbyarsenite163f.
-degradation165-effectonactinomycinbiosynthesis322,326-fermentation,conditions162-,glycosidationoferythronolides166-lactone(erythronolide),biosynthesis163,441--originof165-methylgroupfrommethionine392erythronolideseeerythromycinlactoneetamycin286,298ethionine117ethyl-IX-thiolincosamidine(ETL)354euphanegroup145,148euphol144,145falcarinone441trans-trans-farnesylpyrophosphate135,138f.
fattyacids,branchedchain,inbiosynthe-sisoferythromycin163ferrichrome17,439-,biosynthesis439ferrichromeA,fromt5-N-hydroxyorni-thine439ferrimycin17456SubjectIndexflagella,bacterial,e-N-methyl-lysinein301flavacol23,49flavipin90-degradation91flavomycinseeneomycinfolicanthine437formaldehyde,forD-psicose449formate18,386,440formylglycine18,20N-formyl-p-(methoxy)-styrylamine286-formyl-saIicylicacid85forosamine(2,3,4,6-tetrydeoxY-4-dime-thylamino-hexose154,155,158,173f.
,178ff.
fradiomycinseeneomycinframycetinseeneomycinD-fructose,incorporationintopsicofura-nine403D-fucoseinbiosynthesisofmacrolidesugars179fulvicacid125,127,128fungi,dermatophytic433-,pathogenetic449fumagillin152ff.
-biosyntheticscheme153fumigatin91ff.
fusarubin127,128fuscin99-synthesisfromacetate-polymalonateroute99f.
fusidicacid144ff.
-production145genipicacid137genipin137genipinicacid138gentamycins361gentisaldehyde85gentisicacid85,124gentisylalcohol85geodin96,118,130-biosynthesis97-hydrate97geodoxin97geranylgeranylpyrophosphate135geranylpyrophosphate135-unitinmycophenolicacid1oof.
gladiolicacid90gliotoxin29,439D-glucosamine362,365,368f.
,391D-glucose,formationofN-methyl-L-glu-cosamine389ff.
-,--streptose387ff.
-incorporation39(p-nitrophenylseri-noll,62(pyocyanin),171(magnamy-cins),361f.
,365,368(neomycins),391(streptomycin),401(cordycepose),403(psicofuranineanddecoyinine)L-glucose,notintostreptomycin391D-glucose-6-phosphate394D-glutamicacid361L-glutamicacid218ff.
(variotin),361(neomycin)glutarimideantibiotics222f.
-ringofcycloheximide225f.
glutathione7,286glutinone147,148glycerol57f.
glycine301-incorporation18(hadacidin),342(edeine),378(streptidine),421,424(prodigiosin)439(ferrichrome)glycolyticpathway395glyoxylicacid20gramicidin240f.
,245f.
,246,286-S245,246-effectonproteinsynthesis329f.
grifolin134,138f.
grisans123griseofulvin(2S,6'R)-7-chloro-4,6,2'-trimethoxy-6'-methylgris-2'en-3,4'-dione)96,123ff.
,440-biosyntheticpathway125ff.
(scheme),127---,halogenation131---,methylation130f.
-,degradation127-improvementofyieldbyN-methylcompounds126-production125griseomycins157,175L-gulose390hadacidin17ff.
,439-degradation18helvolicacid134,145f.
-production145f.
heptadecanoicacid163(erythromycin)hexa-N-acetylneomycin364L-histidine361(neomycin)L-homoarginine379(streptidine)homoorsellinicacid(2-ethyl-4,6-dihydro-benzoicacid)85humulene140hydronaphthacenicacidskeleton(tetra-cycline)115hydroxamates17,22f.
hydroxamicacids,biosynthesis4393-hydroxyanthranilicacid(3-HAA)309ff.
hydroxyaspergillicacid43p-hydroxybenzoicacid85-,precursorofAringofnovobiocin236.
SubjectIndex4574-hydroxy-2,2'bipyrrole-5-carboxalde-hyde(HBC)417p-hydroxycinnamicacid,precursorofBringofnovobiocin2357-hydroxycoumarinbiosynthesis23510-hydroxy-2-decenoicacid2925-hydroxy-Sa,11a-dehydrotetracycline120N-hydroxyglycine18,20ff.
,22D-IX-hydroxyisovalericacidinvalinomy-cin2693-hydroxykynurenineinactinomycin309ff.
hydroxylaminoacids(hadacidin)20ff.
2-hydroxy-3-methoxy-5,6-dimethyl-1,4-benzoquinone91f.
3-hydroxy-4-methylanthranilicacid(4-MHAA)inactinomycin3094-hydroxy-3-(3-methylbutyl)-benzoicacidindihydronovobiocin2374-hydroxy-6-methylpretetramid114ff.
hydroxymycinseeparomomycin2-hydroxyparidine311hydroxyphenazine586-N-hydroxyornithine23,439m-hydroxyphenylalanine(m-tyrosine)30hydroxyphthacenicacidskeletonintetracyclines,biosynthesisfromace-tate1153-hydroxyphthalicacid85hydroxY-L-proline286,298ff.
(actinomy-cin)2-hydroxypyridine-N-oxide43hydroxystreptomycin374,389L-hydroxystreptose(3-C-formyl-Iyxo-furanose)3745-hydroxytetracycline35,113,120illudinM140-S139f.
--,production139"Ilotycin"seeerythromycininactone222iridodial136iridomyrmecin136iron,inhibitionofhydroxymatesynthesis24isanicacid1893-isobutyl-6-(1-hydroxY-1-methyl-ethyl)-1-hydroxY-2(1H)pyrazinoneseemuta-aspergillicacid3-isobutyl-6-sec.
-butyl-2-hydroxy-pyrazine-1-oxideseeaspergillicacidisocycloheximide222,223isoiridomyrmecin136isonovobiocin232f.
,233isopenicillinN1,12isopentenylpyrophosphate134,135isophenoxazinesynthetase318isopyrrolnitrin(2,3-dichloro-4-(2'-nitro-phenyl)-pyrrole)434,435f.
isorhodomycinones102,103isotopecompetitiontechnique61isotyrosineinedeine3424-0-isovaleryl-mycaroseinleucomycins175junipal202kanamycinB360f.
5-keto-2,6-diamino-2,6-dideoxy-D-glu-cose,precursorofdiaminohexosefragmentinneomycins361ff.
4-ketodedimethylaminoanhydro-7-chlor-tetracycline114,1184-ketodedimethylaminoanhydrotetra-cycline114,117kojicacid440lachnophyllumester,189,B-lactam-dihydrothiazineringsystem(cephalosporins)12,B-lactam-thiazolidine(6-APA)ringsystem(penicillins)3,10,12lactate39lankanolide(aglyconeoflankamycin)168-biosynthesis169lankamycin156ff.
,168f.
,172,176ff.
D-lankavose(4,6-dideoxy-3-0-methyl-D-xylohexose)154,166,158,168,172,178ff.
lanosterol144,145(fusidicacid),147(eburicoicacid)D-leucine,46(aspergillicacid),261(poly-myxin)-,inhibitorofpolymyxinformation260L-leucine57(pyocyanin),172(L-myca-rose),261(polymyxin)cyclo-leucyl-leucyl23,49leukomycins157ff.
,175limonene136f.
lincocin(R)seelincomycinhydrochloridelincomycin363ff.
-biosynthesis,precursors355--,scheme357f.
-degradation356-hydrazinolysis353f.
-originofmethylgroups356macrolideantibiotics164ff.
--,biosynthesisbyacetate-polypro-pionatepolycondensations103--,-andnystatin228--,-,sugarparticipants158458SubjectIndexmacrolideantibiotics,biosynthesis,summary176f.
--,physicalconstants159--,producers(table)156f.
macrolidesugars155--,tentativepathwayforbiosynthe-sis178ff.
(flowsheet),178macrolides,6-deoxyhexosesattachedto154f.
,155-,oxidationto3-hydroxY-2.
4,6-tri-methylpimelicacidlactone169-,summaryofbuildingunits177magnamycin154ff.
,170,176ff.
-biosynthesisfromglucoseandsucci-nate171-,hydrolysis170-,precursors163malonate101(rugulosin).
127ff.
,440(griseofulvin),224f.
,(cycloheximide),(scheme)226;227(streptimidone)malonyl-CoA89(alternariol),115(tetra-cyclines),199(polyacetylenes)malonyl-pantotheine89(alternariol)maltol(3-hydroxy-2-methyl-4-pyrone),fromstreptose387maltose361(neomycins)D-mannose392(mannosidostreptomycin)mannosidostreptomycin(streptomycinB)375-,synthesis392f.
cx-mannosidostreptomycinase393marasin193-,enzymaticformationofalIenicsystem199marasmicacid140,141"matatabilactone"136matricariaester189,441"matromycin"seeoleandomycinmellein89"metatetrane"118f.
methionine,methyldonorinactinomycin308(table)cycloheximide224f.
fumagillin152lincomycin356N-methyl-L-glucosamine392f.
pyocyanine63streptose386variotin218,220-,precursorofpenicillinN63-methoxy-2,2'-bipyrrole-5-carboxalde-hyde4134-methoxy-2,2'-bipyrrole-5-carbox-aldehyde(MBC),precursorofprodi-giosin412ff.
,450N-methylalanine286N-methylalaninebiosynthesis301(actinomycin)N-methylaminoacidsinantibiotics3012-methyl-3-amylpyrrole(MAP),precur-sorofprodigiosin412ff.
2-methylanthraquinone1-methyl-4-ethyl-L-proline(ethylhygricacid),biosynthesis356N-methyl-L-glucosamine(N-methyl-2-amino-2-desoxy-L-glucopyranose)373f.
,378-,biosynthesis389ff.
-,mechanismofformation390-,penta-O-acetylderivative389methylgroups,migration444methylgroups,origininactinomycin308cycloheximide224f.
fumagillin152lincomycin356N-methyl-L-glucosamine392f.
myomycin329--,pyocyanine63streptose386U-21,699,356variotin218ff.
6-methylheptanoate(MHA)inpoly-myxins2552-methyl-3-heptyl-4-propylpyrrole4134-methyl-3-hydroxyanthranilicacid(4-MHAA)-peptidesinactinomycin310ff.
4-methyl-3-hydroxyanthraniloylpenta-peptideinactinomycinbiosynthesis292N-methyl-isoleucine292methylmalonate,incorporationintoerythromycins1662-methyl-5-methoxybenzoquinone91f.
N-methyl-mitomycinA676-(+)-methyloctanoate(MOA)inpoly-myxins255,262methylorsellinicacid92f.
6-methylpretetramid114ff.
3-methyl-DL-prolineinactinomycinbio-synthesis327ff.
4-methylproline3002-methylpyrrole413,425N-methyl-2.
4-quinazolinedionefromcyclopenol1066-methylsalicylicacid83,85,90,124,130--,acetylogs90--biosynthesis83f.
,88,90,92methyl-cx-thiolincosamidine(MTL)353ff.
methyltriaceticlactone95cx-methyl-DL-valine,inhibitorofpenicillinbiosynthesis296SubjectIndex459N-methyl-L-valineinactinomycin277,283,286f.
,296,301ff.
methymolide(aglyconofmethymycin)160methymycin154ff.
,160ff.
,176ff.
-biosynthesis,propionaterule160f.
mevalonate,incorporationintoeburicoicacid147-,--fumagillin152f.
-,--fuscine100-,--mycophenolicacid100-lactone,incorporationintofumagillin153miamycin157,159mitiromycin66mitomycinsA,B,C66ff.
-,biosynthesis68ff.
-,-,fermentationmedia69-,-,inhibitionbyDL-ethionine70-,-ofmitosanenucleus75-,-,reversalbymethionine71adenosylmethionine72-,effectonactinomycinbiosynthesis322,326-nucleus(mitosane)66mitosanemoietyofmitomycins66,75monoanthraquinones,biosynthesisbypolyacetateroute101.
(scheme),102monomycinseeparomycinmonoterpenoids134f.
cis-trans-muconicacid144mucomycinA143muta-aspergillicacid46D-mycaminose(3,6-diedeoxy-3-dimethyl-amino-D-glucose)154,155,158,170ff.
,174,178ff.
L-mycarose(2,6-dideoxy-3-C-methyl-L-ribohexose)154,155,162,170ff.
,174f.
,178ff.
,441-fromglucose1713-(X-L-mycarosylerythronolide440mycelianamide17,29,104,124f.
,134-degradation105-,formationofaromaticring105mycifradinseeneomycinmycinones103f.
D-mycinose(6-deoxY-2,3-di-O-methyl-D-allose)154,155,158,172ff.
,178ff.
mycobacillin271ff.
-,antifungalspectrum271-biosynthesisandmetabolism272,445--,effectofmedia272--,incorporationofaminoacids273446(table)--,inhibitionbychloramphenicol273--,noninterferenceofstreptomycindependence273mycobacillinbiosynthesis,roleofnucleo-tide-linkedpeptides273,446--,studieswithnon-producermutants273-,production272mycobactin17mycomycin193ff.
mycophenolicacid99--,derivationofsidechain100--,synthesisfromacetatepolymalo-nateroute99f.
mycosamineinnystatin228myo-inositolinstreptidine382ff.
,390---streptomycin447myo-inosose,inhibitionofstreptomycinproduction385myomycin392naramycinAseecycloheximide222-B222,223narbomycin156ff.
,169f.
,176ff.
-biogeneticrelationshiptoothermacrolides169neamine359ff.
nemotin192nemotinicacid192neoaspergillicacid23,48-production48neobiosamine359,365neohydroxyaspergillicacid48-production48neomethymycin155f.
,159,160,176ff.
neomycinA359ff.
-B359f.
-C359ff.
neomycins,biosynthesis,relationtocellwallsynthesis370-,degradationofsubunits(scheme)366-,effectonactinomycinbiosynthesis322,326-,precursors(table)363-,productionfromaminoacids361-,--sugars361-,subunits,biogeneticscheme362neosamineB359ff.
--labellingfromprecursors(table)365-C359,361f.
,364f.
,368,370--labellingfromprecursors(table)365,368neosamines,degradation365ff.
neospiramycins174neutramycin157,159,176niddamycin157,170f.
176ff.
o-nitroanisol311o-nitrobenzoicacid311nitrogroupinarylring,derivation40460SubjectIndexD-p-nitrophenylserinol(pNPS)inchlor-amphenicol35ff.
norprodigiosin413,415norvaline,inhibitoroftyrothricinforma-tion249noviose(Cringofnovobiocin)fromglu-cose231ff.
(scheme),233-biosynthesis,involvementofUDP-sugars232f.
vitaminB12232f.
novobiocicacid(B+Cringofnovobio-cin)231--,formation(scheme)236--,ringcoupling,enzymespecificity236novobiocin231f.
-biosynthesisofAring235f.
---Bring(coumarinmoiety)234f.
---Cringseenoviose--effectofprecursorsonrate235f.
--energyrequiringcouplingofringsystemsbycell-freeextracts236f.
--,proposedscheme237nystatin228ff.
-biosynthesisfromacetateandpropio-nate228ff.
-degradationtotiglicaldehyde228f.
-labellingpattern228f.
nystatinolide(aglyconofnystatin)228odyssicacid200oenanthotoxin194"Oleandocyn"seeoleandomycinoleandolide(aglyconeofoleandomycin)167f.
oleandomycin156ff.
,167ff.
,176ff.
-biogeneticscheme168-productionconditions168-triacetate(TAO)167L-oleandrose(2,6-dideoxY-2-0-methyl-L-arabohexose)154,155,158,167,178ff.
ommochrome(phenoxazinone)synthesis317oospolactone(C-methylderivativeofmellein)89orcinol(1,3-dihydroxY-4,5-dimethyl-benzene)91ff.
,138orcylaldehydeinstipitaticacid94DL-ornithineinprodigiosin421orsellinicacid82f.
,85,88,90,93,125,126,1384-oxo-6-deoxy-hexose,keyintermediateoferythromycinsugars1674-oxo-L-proline286-biosynthesis(actinomycin)298ff.
--distributionoflabel300oxygeninhydroxylaminogroup22oxypyrrolnitrin(3-chloro-4-(2'-nitro-3'-chloro-6'-hydroxy-phenyl)-pyrrole)434oxytetracyclinesee5-hydroxytetracyclinepalitanin128paromycins360,361patulin85,86,88,90,124D-penicillamine(D-p-thiovaline)4,296-,incorporationofvalines296penicillicacid85ff.
,87,88ff.
penicillins1ff.
-,aminoacidprecursors3ff.
-,chemicaltransformationtocephalo-sporins10-,6-APA-ringprecursors3f.
-,intermediates7f.
-,sidechainprecursors3f.
penicillinN1f.
,3ff.
-configurationofpenicillicacid296-effectonactinomycinbiosynthesis322,326pentadecanoicacidinerythromycins1632-pentenylpenicillinR1pentoseshuntinstreptosesynthesis387,395phenoxazinesynthesis,oxygenuptake316phenoxazinonesbyoxidativecondensa-tionfromo-aminophenols3174-MHAA-peptides315phenoxazinonesynthetase31Off.
--,inhibitionbyaminobenzenederi-vatives312ff.
chloramphenicol(table)3263-hydroxyanthranilicacidderivatives311ff.
--,metaleffects315f.
--,substratespecificity310phenoxy-methylphenylpenicillinmethylestersulphoxide10phenylalanine,precursorofchlorampheni-col35f.
-,--gliotoxin30-,--tropolones93-,--viridicatin106phenyl-et-mannosidehydrolysis393phenylpyrrolederivatives433ff.
---,halogenated434f.
phosphatidylethanolamine302phosphoenolpyruvate39o-phthaldialdehydes90f.
physostigmine437phytoene135picromycin154ff.
,155,158,176ff.
-biosynthesisseemethymycine159SubjectIndex461CSa-pigment(prodigiosin)413piperidinecarboxylicacidinactinomycinbiosynthesis288f.
,290f.
polyacetylenes189f.
,441f.
-,biogeneticcapabilities208f.
,209-,biosynthesis,feedingexperiments206ff.
-,cyclizationreactions203ff.
-,epoxide203f.
-,formationbydehydrogenation196-,furane203ff.
-,introductionoffunctionalgroups200.
-,mechanismoftriplebondformation195f.
-,originofC-skeleton199f.
-,spiroketalsof205,443-,thioether203ff.
poly-,B-ketides82ff.
,98,B-polyketomethylenechainfromacetate126ff.
--ingriseofulvinsynthesis127ff.
polymyxins254,255-,biosynthesis321-,-,inhibitors260ff.
-,-,mechanism260.
-,-andproteinsynthesis260-,--sporulation263f.
-,compositionof(table)254-,producingstrains255,(table)256-,production,media256f.
,258f.
-,-,carbonsource(table)256-,-,and2,4-diaminobutyricacid263-,-,nitrogensource(table)257-,-,andpH259-,-,andgrowthpolyphenoloxidase317porfiromycin66pretetramid116f.
pristimerin134,147f.
prodigiosin410ff.
,412,449f.
-biosynthesis414ff.
,(scheme)415--,aminoacidrequirement418--,isotopestudies420--,andthiamine420-,biosyntheticanalogs413f.
--,condensationreaction425--,environmentalfactors421-,chemistry411ff.
-,functions426-,fungicidalaction411-production,media418--,effectofair421f.
metals419f.
phosphate420temperature421f.
--,inhibitionbyantibiotics424-,sclerosingeffectinveins411prodigiosinsecondarymetaboliteinagedcultures426-,syntheticanalogs413-,water-solubleforms414-,cultureconditions450prodigiosin-likepigments,producedbyactinomycetes413L-prolineintoactinomycin277,291,298f.
,300-intoprodigiosin421ff.
propiolicacid189,195propionicacidintomacrolides160ff.
,163propylhygricacid(PHA)(trans-1-methyl-4-propyl-L-proline)353ff.
propylsuccinicacidfromlincomycin356proteinsynthesisandactinomycinforma-tion320ff.
---edeineformation346ff.
protetrone116protomycin222protozoa,inhibitionbyprodigiosin410psicofuranine(angustmycinC,adenine-alluloside)400,403ff.
,448f.
-biosynthesis403,449n-psicose(n-allulose)403-fromhexoses403-phosphate(n-allulose-6-phosphate)449puberulicacid93f.
puberulonicacid93f.
pulcherrimin17pulcherriminicacid23,50puromycin,effectonactinomycinbio-synthesis322,326-,--bacitracinbiosynthesis244-,--edeineproduction345f.
-,--tyrothricineformation249pyocyanine52-,carbonsources57,62-,degradation59-,productionbycellsuspensions56-,--growingcultures52-,-,media55pyrogallol85pyrrole425pyrrolnitrin(3-chloro-4-(2'-nitro-3'-chlorophenyl)-pyrrole)(=antibioticA10338)433f.
-antimicrobialspectrum433-incorporationoflabelledcompounds436f.
,437(table)-isolation434-mediumforproduction435-pathway437-precursors435f.
,-tradename:PYRO-ACE462SubjectIndexpyrromycinones102f.
,103pyrryldipyrrylmethene411ff.
pyruvateinpyocycaninesynthesis58--griseofulvin440pyoluteorin434quadrifidins193quadrilineatin90quinicacidinpyocyanin62o-quinonimineinphenoxazinesynthesis317D-quinovoseinmacrolidesugars179ramycinseefusidicacidrelomycin157,174f.
,176ff.
-fromtylosinbyreduction175ribonuclease,exogenous445ribose359,365(inneomycin),402(incordycepose)-,degradation366ff.
-,precursors(table)365,369D-ribose-5-phosphate,forD-psicose449ribosomes,bindingofedeine344RNAtemplate445"Romicil"seeoleandomycinroridins143rugulosin101-,C-C-couplingbyradicalmechanism101-,formationfromacetateandmalonate101saccharopine6sangivamycin407sarcosine277,287-biosynthesis301ff.
scyllo-inosamine,phosphateesters447scyllo-inositol(streptidine)384ff.
,390serine18,63,386,439f.
sepedonin95sesquiterpenoids134f.
siderochrome17shikimicacid37,60--pathway29,38,85,93,107,124spermidine344spiramycins157ff.
,173f.
,176ff.
spirodienoneintermediateinnovobiocinbiosynthesis234f.
,235all-trans-squalene145,147f.
steroids134f.
stipitaticacid94ff.
streptamine(1,3-diamino-2,4,5,6-tetra-hydroxycyclohexane)382,447-phosphateesters447streptidine(1,3-diguanidino-2,4,5,6-tetrahydroxycyclohexane)373f.
,378streptidinebiosynthesis378ff.
,447--,reactionsequence384ff.
streptimidone222,227streptobiosamine373f,388f.
streptomycin373ff.
,447f.
-biosynthesis,C-backbonefromglucose(table)383,(table)387,(table)388frommyo-inositol(table)383--,andgeneralmetabolism394f.
--,pathwaysinS.
griseus393--,sequentialsteps393ff.
-effectonactinomycinsynthesis322,326---mycobacillinsynthesis273---prodigiosinsynthesis424-,productionmedia376-,roleinlifeofproducers395L-streptose(3-C-formyl-5-deoXY-L-lyxo-furanose)373f.
,386-fromglucose386ff.
,448-,degradation388-,mechanismofformation387ff.
streptothricinBIseeneomycin-BlIseeneomycinstreptovitacinA,BandC.
222succinateintomagnamycins171--prodigiosin421,424N-succinyl-D-valine295f.
sulochrin97,124,129,130synthrophicpigmentationinS.
mar-cescens416tatiricacid189telomycin298terpenoidantibiotics134ff.
terphenylquinone30terthienyls444tertiomycins176a-terthienyl202tetracyclines113-,biosyntheticpathway(scheme)114-,effectonactinomycinbiosynthesis322,326-,-bacitracinbiosynthesis244-,--prodigiosinbiosynthesis424-,hydronaphthacenicacidskeleton115thioethers,polyacetylenic201.
,443f.
thiophenes,inpolyacetylenesbio-synthesis443f.
L-threonine277,283,293thymidinediphosphate-glucose(TDP-glucose)388,393--mannose(TDP-mannose)393--rhamnose(TDP-rhamnose)388SubjectIndex463tiglicaldehydefromnystatin228tirucal1anegroupofterpenoidanti-biotics145,148tirucallol144,145toyocamycin(cyano-7-deazaadenine-ribonucleoside)400,405,407-,adenineasprecursos407transamidinasesinstreptomycinproduction380f.
,447tricarboxylicacidcycleinstrepto-mycinformation395trichodermin142f.
trichodermol142f.
trichothecin141ff.
-production1423,5,7-trioxooctanoicacid126tripyrrylmethene411triterpenoids134f.
tropolones93ff.
trypacidin96n-tryptophan435f.
L-tryptophanintomitomycin75--violaceine78tryptophanmetabolism433tubercidin(4-aminopyrrolo-(2,3d)-pyrimidine-fJ-n-ribofuranoside)400f.
,405f.
,448tuberculostearicacid171tylosin157.
,174f.
,176ff.
-,reductiontorelomycin175tyrocidine240f.
,245f.
-,effectonproteinsynthesis329f.
L-tyrosineintomitomycin75--novobiocin234f.
--tropolones93--xanthocillin27f.
tyrothricin(mixtureofgramicidinandtyrocidine)247f.
-biosynthesis,conditions247.
--,inhibition250--,mechanism249f.
--,andproteinsynthesis250ubiquinone91f.
uridinediphosphate-N-acetylglucos-amine(UDP-NAG)inneomycinbiosynthesis369f.
usticacid89n-valineintoactinomycin277,293ff.
,296--valinomycin269-,utilizationbyS.
antibioticus293ff.
L-valine,conversionton-valine(actinomycinbiosynthesis)296-,intovalinomycin269-,utilizationinactinomycinbio-synthesis(table)297)valinomycin268ff.
-precursors269vanomycin,effectonactinomycinbiosynthesis322,326variotin216ff.
,217-biogenesisfromacetate217,218,220--pyrrolidonemoiety219-degradation216ff.
,(flowsheet)217-mediaforproduction217verrucarin143ff.
-Af42f.
,143-B143-,biogeneticscheme144-production143verrucarol143,144violacein77ff.
,450-,antibioticproperties79-,metabolicfunction78-,physiologicalrole78-,radiationprotection78-,toxiceffects79-,tryptophanin78viridicatin(2,3-dihydroXY-4-phenyl-quinoline)30,105f.
-biosynthesis,hypotheticalmecha-nism106f.
viridicatol(3'-hydroxyviridicatin)106f.
volucrisporin30xanthocillin26ff.
-,degradation27-,N-formyltoisonitrilegroups28-,precursors27xanthommatin317xanthones127,129ximenynicacid195zygomycinAseeparomomycinIndexofOrganismsActinidiapolygamaMIQ.
136Actinomycesscabies32Aerobacteraerogenes404,449Agaricaceae190Alcaligenesmetalcaligenes32Alternariatenuis89Anaphalis442Anthemissp.
201.
,204,443-tinctoria208Araliaceae190ArtemisiaannuaL.
202,207-capillaris193-vulgaris442Aspergillussp.
29-candidus98-Ilavus(PRL)43ff.
-fumigatusFRES.
83,92,96,152--mut.
helvola145-melleus89-niger433-oryzae45f.
-sclerotiorum(strainNRRL415)48ff.
-terreusTHOM.
93,96,98,130Bacillusanthracis286-brevis245,(Vm4)342ff.
,446-colistinus255,257-lichenilormis240-prodigiosus,seeSerratiamarcescens-pyocyaneus(Pseudomonasaeruginosa)52ff.
-subtilis79,271,289,364,433-polymyxa255ff.
Bacteriumprodigiosum,seeSerratiamarcescensBasidiomycetes190,192BidensconnatusMUHLENBG.
443BuphthalumsaliciloliumL.
444Blastomycesdermatidis216,449Caldariomyceslumago98Calycanthaceae437Calycanthusoccidentalis385,437Candidaalbicans433Carlinaacaulis204,208Celastrusdispermus(Maytenusdisper-mus)147-paniculatus147Celastrusscandens147-strigillosusNAKAI147CentaurearuthenicaLAM.
196,202,206Cephalosporiumsp.
1ff.
,146-lamellaecola144-mycophyllumIFO6615145ChaetomiumcochloidesPALL.
83f.
-globosum83ChamaemelumnobileL.
443ChimonanthusLINDL.
437ChromobacteriumprodigiosumseeSerratiamarcescens-violaceum78ff.
Chrysanthemummaximum200-segetumL.
207-serotinumL.
442f.
Cicutavirosa194Cladosporiumlulvum83Clitocybediatreta190,192-illudens,strain72027-5139f.
Clostridiumleseri32-welchii79Coccidioidesimmitis411Compositae190Coprinusquadrilidus190,193,200Cordycepsmilitaris401,448Coreopsissp.
203Cosmossulphureus200CousiniahystrixC.
A.
MAY200Curvularialunata83Daedaleajuniperinus202Dahliasp.
442Denhamiapitoosporoides147Drosophilasubatrata192Echinopssphaerocephalus208,444Entamoebahistolytica411Entoloma190Emericellopsissp.
1Escherichiacoli39,189,324,433Euphorbiaceae190Fomesol/icinalis146Fusidiumcoccineum144GenipaamericanaL.
137f.
Gibberellalujikuroi83IndexofOrganisms465Gliocladiumsp.
29-roseumBAINIER91Gnaphalium442Gramineae190Grifoliaconfluens138Helminthosporium448Hippocrateaindica147Histoplasmacapsulatum449Hydnum190lchthyothereterminalis194Iridomyrmexhumilis136Lactobacillusleichmanni402Lampteromycesjaponicus(KAWAM.
)SINGH139Lauraceae190Lentinusdactyloides146-degener91Leucopaxillusalbissimus191Loranthaceae190Marasmiuscongenus(Mo.
6890)140-ramealis193MicrococcusprodigiosusseeSerratiamarcescensMicrosporumaudouini216Minoliales190Mucorramannianus145Mycena190Mycobacteriumtuberculosis433MyrotheciumroridumS1135143-verrucaria143Neisseriameningitidis79Nitrobacteragilis21Nocardiaacidophila190-carallina175Oenanthecrocata193OidiodendronfuscumROBAK101Olacaceae190Opiliaceae190Oosporasp.
90-sulphurea-ochracea129Paecilomycespersicinus1-variotiBAINERvar.
antibioticus216f.
Papilionaceae190PastinacasativaL.
441Penicilliumsp.
29-albidumSoPP.
C.
M.
I.
40219124-aurantio-violaceum18ff.
--virensBIOURGE93Penicilliumbaarnense83f.
,103-brefeldianumDODGE124-brevicopactum83,90,101-brunneumUDAGAWA101-chrysogenumHf.
,433-citrinum98-cyclopiumWESTLING(NRRL1888)83,86,94,105-"estinogenum"97-frequentans99,128-gladioli100-griseofulvumDIERCKX83,104,124ff.
,440-islandicumSopp101-janczewskiiZAL.
[Ponigricans(BAI-NIER)THOM]124-janthinellum124-madriti83-meliniiTHIM.
124-notatumWESTLING26f.
-patulum(syn.
P.
urticae)BAIN83,85ff.
,124ff.
,440-paxilliivar.
echinulatum130-raciborskiiZAL.
124-raistrickiiSMITH124-rugulosumTHOM.
101-stipitatum83,93,95-urticae(=P.
patulum)125-viridicatumWESTLING105,107PhysostigmavenenosumBALF.
437Phytophthoracinnamoni191Polyporaceae190Polyporusanthracophilus146,199-biformis192-hisPidus146-sulphureus146-versicolor129Poriasp.
192-cocos146Pseudomonassp.
197-aeruginosa52f.
,434-aureojaciens434,436-bromoutilis434-methanica449-pyocyaneaseePs.
aeruginosa-pyrrociniasp.
n.
433Pristimeragrahami147-indica147Proteusvulgaris433Pycnoporussuccineus318Rhodotorulaglutinis191Rickettsiae32Saccharomycescerevisiae79Santalaceae190,195Santalumacuminatum199466IndexofOrganismsSerratiamarcescens410f.
,416,422,424,450--,mutant9-3-3412ff.
--,prodigiosin,nonproducermutants415ff.
--,sponteanouscolormutants415f.
--,temperature-inducedpigmenta-tion422,424Simarubaceae190Staphylococcusaureus289,324Streptomycessp.
1,102,154,224,269,279ff.
,316-albireticuli156,157,170,176-albus155,224-amakusaensis28-ambofaciens157,173-antibioticus156,157,167f.
,276ff.
-ardussp.
n.
66,67-aureofaciens115ff.
,157,281ff.
,285-aureoverticillatus450-bikiniensis157,172,447-bluensis374,447-caespitosus66f.
-chrysomallus277-cirratus157,175-djakartensis157,170-erythreus156,161ff.
,164ff.
,440-eurocidicus157,160,176.
-eurythermus157,175-felleus155ff.
-flaveolus285-flavochromogenes155f.
-flavus285-flavus-parvus285-fradiae157,174,281ff.
,359ff.
-fulvissimus268-griseocarnosus374-griseoflavus155f.
,157,175-griseolus157,175-griseus35,224,373ff.
,447f.
-halstedii156,164-hygroscopicus115,157,174,400,449--var.
angustmyceticus403f.
--var.
decoyicus403-lavendulae157,175-lincolnensisvar.
lincolnensis353ff.
-longisporusruber450-longissimus450-kitasatoensis157,175Streptomycesmacrosporeus159-melanochromogenes281ff.
-michiganensis285-naraensis224-narbonensis156,169-niveus231ff.
-noursei223,228-olivaceus156-parvulus279ff.
-pheochromogenes157--var.
chloromyceticus32-pulveraceus224-reticuli156.
,175--var.
protomycinus224-rimosusf.
paromomycinus224-rimosus35,115ff.
-ruber450-subrutilis374-tendae156-thermotolerans156,170-thioluteus21-tsusimaensis269-tubercidicus405-venezuelae32ff.
,155ff.
-verticillatus66,68ff.
-violaceoniger156,168Tagetessp.
194-erecta202-tenuifoliaCAV.
208Torulautilis433Trichodermaviride29f.
,143Tricholomagrammopodium199f.
Trichophytonasteroides433-interdigitale433-rubrum216,433Trichotheciumroseum141f.
TripterygiumregeliiSPRAGUEetTAKEDA147-wilfordiiHOOK147Trypanosomabrucei411-equiperdum411Umbelliferae190Ustilagosphaerogena439Valerianaceae190ViciafabaL.
191
- methoxychrome相关文档
- 侦测chrome18
- 大连市驾管处科目二考试弱电及场地改造工程
- Surveychrome18
- 测温chrome18
- www.novell.com/documentation
- interposechrome18
酷锐云香港(19元/月) ,美国1核2G 19元/月,日本独立物理机,
酷锐云是一家2019年开业的国人主机商家,商家为企业运营,主要销售主VPS服务器,提供挂机宝和云服务器,机房有美国CERA、中国香港安畅和电信,CERA为CN2 GIA线路,提供单机10G+天机盾防御,提供美国原生IP,支持媒体流解锁,商家的套餐价格非常美丽,CERA机房月付20元起,香港安畅机房10M带宽月付25元,有需要的朋友可以入手试试。酷锐云自开业以来一直有着良好的产品稳定性及服务态度,支...

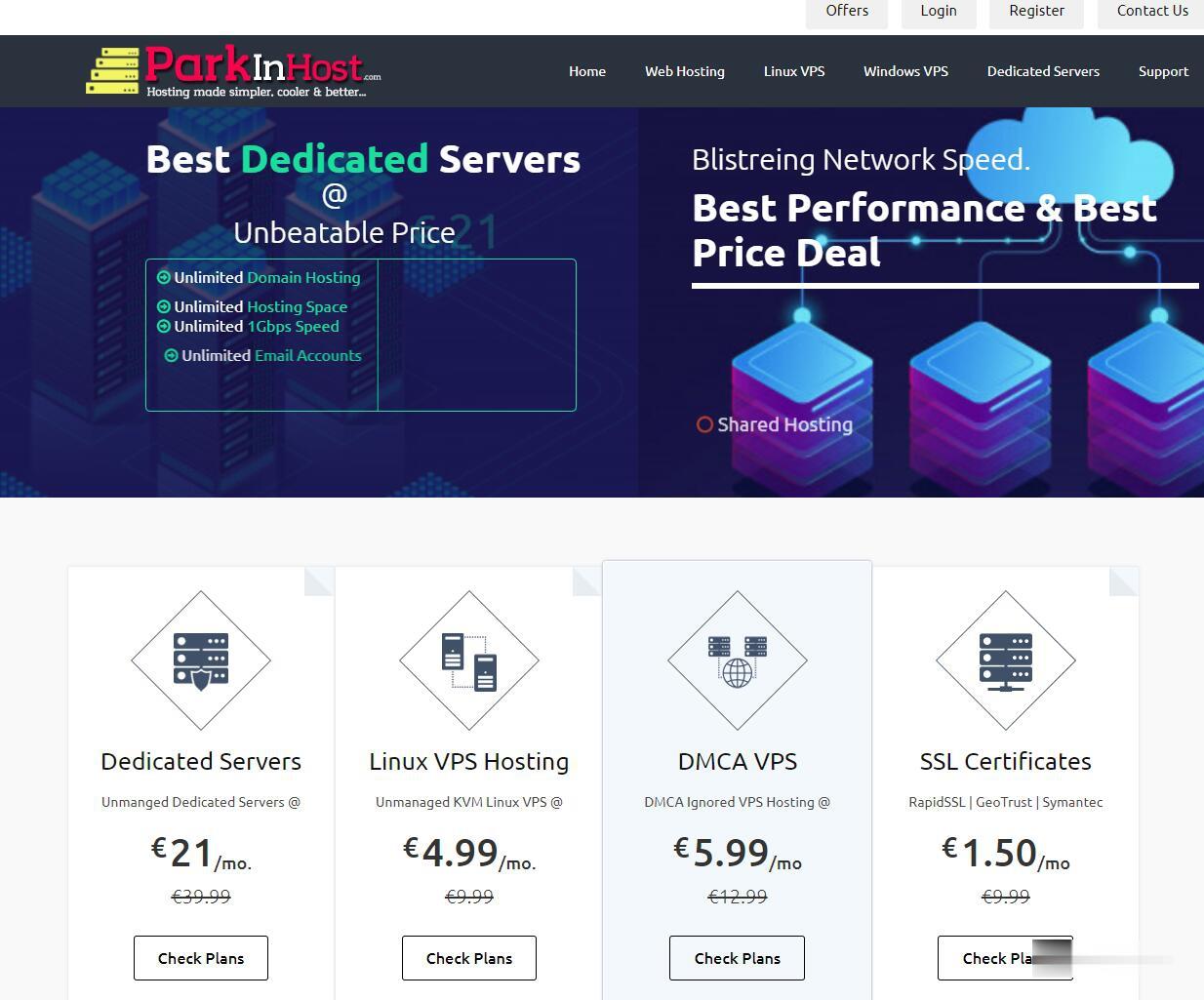
ParkinHost:俄罗斯离岸主机,抗投诉VPS,200Mbps带宽/莫斯科CN2线路/不限流量/无视DMCA/55折促销26.4欧元 /年起
外贸主机哪家好?抗投诉VPS哪家好?无视DMCA。ParkinHost今年还没有搞过促销,这次parkinhost俄罗斯机房上新服务器,母机采用2个E5-2680v3处理器、128G内存、RAID10硬盘、2Gbps上行线路。具体到VPS全部200Mbps带宽,除了最便宜的套餐限制流量之外,其他的全部是无限流量VPS。ParkinHost,成立于 2013 年,印度主机商,隶属于 DiggDigi...
百驰云(19/月),高性能服务器,香港三网CN2 2核2G 10M 国内、香港、美国、日本、VPS、物理机、站群全站7.5折,无理由退换,IP免费换!
百驰云成立于2017年,是一家新国人IDC商家,且正规持证IDC/ISP/CDN,商家主要提供数据中心基础服务、互联网业务解决方案,及专属服务器租用、云服务器、云虚拟主机、专属服务器托管、带宽租用等产品和服务。百驰云提供源自大陆、香港、韩国和美国等地骨干级机房优质资源,包括BGP国际多线网络,CN2点对点直连带宽以及国际顶尖品牌硬件。专注为个人开发者用户,中小型,大型企业用户提供一站式核心网络云端...

chrome18为你推荐
-
上海fastreport2I:\Sam-research\QEF\Publications\Conference更新win7点击ipad支持ipad支持ipadipad连不上wifi苹果ipad突然连不上网了,是怎么回事?网络是好的,手机能上网。联通版iphone4s苹果4s是联通版,或移动版,或全网通如何知道?360chromechrome是什么文件夹?是360急速浏览器吗?但是怎么没有卸载掉?谷歌sbSb是什么意思?