genesis777k7.com
777k7.com 时间:2021-03-18 阅读:()
RESEARCHOpenAccessSmoothenedgenealterationsinkeratocysticodontogenictumorsZhangRui1,PengLi-Ying1,QuJia-Fei1,HongYing-Ying1,ChenFeng2*andLiTie-Jun1*AbstractBackground:Ithasbeenwidelydemonstratedthatthehedgehogpathwayisstronglyassociatedwithbasalcellcarcinomaoftheskin(NBCCS).
ToassesspotentialDNAalterationsrelatedtokeratocysticodontogenictumors(KCOTs),wesequencedsmoothened(SMO)genesin12sporadicKCOTs.
Methods:Polymerasechainreaction(PCR),capillaryelectrophoresisanddideoxychain-terminationsequencingwereusedtoexaminepotentialDNAalterationsinsporadicKCOTs.
Results:FivealterationsinSMOgenesweredetected.
Fourofthesemutationsconsistedoftwosynonymousandthreemissensemutations;twoofwhichhavenotbeenreportedtodate(c.
T776A,c.
T1281G).
Conclusions:SMOgenesmayplayanimportantroleinthesonichedgehog(SHH)pathwayandcouldalsoberesponsibleforgeneratingKCOTsandNBCCS.
However,theirinfluenceonSHHsignalingremainstobeelucidated.
Keywords:Keratocysticodontogenictumor(KCOT),Hedgehog(HH)signalingpathway,Genemutation,SMOgeneBackgroundOdontogenickeratocyst(OKC)isanaggressive,cysticjawlesionwithstronggrowthpotentialandahighrecurrencerate.
Inrecentyears,theWorldHealthOrganization(WHO)reviseditsnametokeratocysticodontogenictumor(KCOT).
Thisreclassificationisbasedonitsaggressivebehaviorandhighrecurrencerate,emphasizingthatKCOTisabenigntumorratherthanacyst[1-3].
Althoughthegreatmajorityofkerato-cystsoccurinisolationassingle,non-syndromiccysts,theymayalsopresentasmultiplecystsasafeatureofthenevoidbasalcellcarcinomasyndrome(Gorlinsyndrome,OMIM#109400)[4].
Atpresent,therearemanymanuscriptsthatfocusontherelationshipbetweenKCOTandPTCH1(patched)genemutations,demonstratingthatPTCH1,thegeneresponsibleforNBCCS,mayalsoplayanimportantroleinsporadicKCOTs[5-8].
ThePTCH1geneisatumorsuppressorgenelocatedat9q22.
32[9].
Astudyof14patientswithNBCCS-associatedKCOTsand29patientswithsporadicKCOTsindicatedthatmutationsintrans-membrane2(TM2)arecloselyrelatedtothedevelopmentofsporadicKCOTs[5].
Thehedgehog(HH)signalingpathwayisakeyregulatorofembryonicdevelopment,controllingbothcellularpro-liferationandcellfate.
Bindingofsonichedgehog(SHH)toitsreceptor,patched(PTCH1),isbelievedtorelievenormalinhibitionbyPTCH1ofsmoothened(SMO),aseven-spantransmembraneproteinwithhomologytoaG-protein-coupledreceptor[10].
SMOisatumor-relatedgenelocatedat7q32.
3,con-tains12exonsspanningapproximately24kb,anden-codesa787-amino-acidtransmembraneglycoprotein[11].
ItsreceptorisaGprotein-coupledreceptorthatinteractswithPatched,animportantpartoftheHHsignalingpathwayduringembryogenesisaswellasadult-hood[12,13].
TheHHpathwayhasbeendemonstratedtoplayanimportantroleindifferentdevelopment-relatedcancers[14-18],buttheexactmechanismofactionhasnotyetbeenelucidated.
TheproteingeneratedbySMOisdownstreamofPTCH1;thatis,theexpressionofPTCH1restrainstheactivationofSMO,andtherebyinhibitsactivationoftheHHpathway[19-22].
RecentstudieshavehighlightedthetherapeuticvalueofSMO*Correspondence:moleculecf@gmail.
com;litiejun22@vip.
sina.
comEqualcontributors2CentralLaboratory,PekingUniversitySchoolandHospitalofStomatology,22ZhongguancunAvenueSouth,HaidianDistrict,Beijing100081,China1DepartmentofOralPathology,PekingUniversitySchoolandHospitalofStomatology,22ZhongguancunAvenueSouth,HaidianDistrict,Beijing100081,ChinaHEAD&FACEMEDICINE2014Ruietal.
;licenseeBioMedCentralLtd.
ThisisanOpenAccessarticledistributedunderthetermsoftheCreativeCommonsAttributionLicense(http://creativecommons.
org/licenses/by/4.
0),whichpermitsunrestricteduse,distribution,andreproductioninanymedium,providedtheoriginalworkisproperlycredited.
TheCreativeCommonsPublicDomainDedicationwaiver(http://creativecommons.
org/publicdomain/zero/1.
0/)appliestothedatamadeavailableinthisarticle,unlessotherwisestated.
Ruietal.
Head&FaceMedicine2014,10:36http://www.
head-face-med.
com/content/10/1/36antagonistsforthetreatmentofHH-linkedcancers[22,23].
SMO,themainactivatoroftheHHpathwaymayserveasacatalystduringthegenerationofcysts,andtherefore,geneticmutationsofSMOareofgreatimportance.
MethodsTumorsamplesandclinicalbackgroundFourteenKCOTsampleswithadefinitediagnosiswereacquiredfromclinicalsourcesatPekingUniversitySchoolofStomatology,OralandMaxillofacialSurgeryDepartment.
DiagnoseswerebasedonWHOclassifica-tionoftumors:pathologyandgeneticsoftumorsoftheheadandneck[24].
AllsampleswerefromChinesepa-tients(eightmalesandsixfemales).
Agesvariedfrom10to58years,withanaverageof29.
2years.
ExperimentalprotocolsusedinthisstudywerereviewedandapprovedbytheEthicsCommitteeofthePekingUniversityHealthScienceCenter(Peking,China).
Informedconsentwasobtainedfromallsubjects.
DNAisolationandmutationanalysisGenomicDNAsfromepithelialcellsandinterstitialcellsoftumorswereisolatedusingaQIAampDNAMinikit(Qiagen,Hilden,American)accordingtothemanu-facturer'sinstructions.
DNAwasquantifiedusingaNanodrop(ThermoFischer,Wilmington,DE).
SMO(NM_005631.
4)codingsequencesweredeterminedusingpolymerasechainreaction(PCR)and1%agarosegelelectrophoresis.
ThirteenpairsofprimerscoveringtheentirecodingsequencesandafewnucleotidesintotheintronsequencesonbothendsofSMOarede-scribedinTable1.
PCRswereperformedonaPTC-100(MJResearch,Watertown,MA,USA)inafinalvolumeof30μLcontainingapproximately100-ngtemplateDNA,200μmol/LdNTPs,0.
2–0.
4μmol/Leachprimer,1.
25UExTaqpolymerase(TaKaRa,Dalian,China),and10XPCRbuffer(50mmol/LKCl,10mmol/LTris–HCl,pH9.
0,and1.
5mmol/LMgCl2).
Exon1,exon12aandexon12boftheSMOgenewereamplifiedbyatouch-down(TD)PCRtechnique(Table1).
Forexample,TD65-56°Cwasperformedasfollows:aninitialdenaturationstepat95°Cfor5min,followedby40cyclesat94°Cfor30s,30satanannealingtemperaturedecreasingfrom65°Cto56°C(decreaseof1°Ceach2cycles,20cyclesat55°C),30sat72°C,andafinalextensionstepat72°Cfor10min.
Thespecificityofamplifiedproductswasconfirmedby2%agarosegelelectrophoresis.
PCRproductswerepurifiedbyLifeTechnologies(AppliedBiosystems,USA)andsequencedbyanABI3730xlDNAsequencer(AppliedBiosystems,USA).
Bothforwardandreversestrandsweresequenced.
Anymutationde-tectedwasconfirmedusingbothforwardandreverseprimers.
StatisticalanalysisExperimentaldatawereanalyzedusingtheSPSSver.
10.
0softwareandarepresentedasmeans±standarddeviation(SD)usingstatisticalmethodssuchasat-test,analysisofbivariatecorrelation,etc.
APvalue<0.
05wasconsideredtoindicatestatisticalsignificance.
Resultsarerepresenta-tiveoftwoindependentexperiments.
ResultsanddiscussionHedgehog(HH)signalingpathwayHedgehog(HH)isasignaltransductionpathwaycloselyrelatedtocellgrowthanddifferentiation,andplaysavitalroleinembryonicdevelopment.
MutationorabnormalTable1PCRconditionsofSMOexonsExonSize(bp)Forwardprimersequence(5′to3′)Reverseprimersequence(5′to3′)ReferenceAnnealingtemperature(°C)1391GGGCTGCTGCTGCTGCTGGTCCCCGCCCTCTCCAAAACTSunetal.
,2008[25]TD60~502283AAGAACTGTCCTGCCCAGATGCCACTGGACCCTGCCCTATACWangetal.
,2013[12]633305AATAATTTGCCAAGCCAGCCCTTCTGATCATGACCCTTCCCWangetal.
,2013[12]634256AGGGTCATGATCAGAAGGGTCAGTATGCAGTAGGGCAGAGCCWangetal.
,2013[12]635302CTGACTTCTGGGAACCTCCAGGACAGAAGGTGGGTTACTGGCWangetal.
,2013[12]636206GTGGCGCAGGTATAGTGACTGGCCCTATAGGAGCTAGCTGGGWangetal.
,2013[12]637175GACTCCAGAGCCTTAGGACCCTCCCTATGGCTAACTTGTCCCWangetal.
,2013[12]638196AAGCAGTTCTTGGACTGAGCCCCATCCATTGAATCTGCTGTCWangetal.
,2013[12]559379AGTTGGAAGCTGCAGTGGGCAAGGCTGTGCTAGAGGCAGWangetal.
,2013[12]6310228CTCTGGAAAGAATGGCATCGTTCCAAATAATCTGTGTGCCCWangetal.
,2013[12]5511215AATGGCACTGACTATGGGAGGCCACTCTTCAGATCCTCTGGGWangetal.
,2013[12]6312a292GAGCCAGGGCCCCAGGCTCGTACGCTCCCTGTCGGCAAGAGTSunetal.
,2008[25]TD65-5612b315AGTACCATTCCTCGACTGCCTGGTATTGGTTCCTCTCTTTCCSunetal.
,2008[25]TD65-56Ruietal.
Head&FaceMedicine2014,10:36Page2of7http://www.
head-face-med.
com/content/10/1/36expressionofcomponentsofthispathwaywillleadtovariousdevelopmentaldefectsand/ortumors.
AlteredHHsignalingisimplicatedinthedevelopmentofapproxi-mately20-25%ofallcancers,especiallythoseofsofttissues[26].
ThesefindingsalsosuggestthatproteinsoftheHHsignalingpathwayarepredominantlylocatedwithintheepithelialcomponentsofglandularodontogeniccysts(GOCs)anddentigerouscysts(DCs).
Therefore,theHHsignalingpathwaymayplayanimportantroleintheformationofepitheliallining[27].
ThediscoveryofoncogenicmutationsintheHHandmitogen-activatedproteinkinase(MAPK)pathwaysinover80%ofame-loblastomas,locallydestructiveodontogenictumorsofthejaw,wasreportedin2014bygenomicanalysisofarchivalmaterial.
MutationsinSMOarecommoninameloblastomasofthemaxilla,whereasBRAFmuta-tionsarepredominantintumorsofthemandible[28].
Inaddition,SHHsubgroupmedulloblastomasaregen-eticallydistinctininfants,childrenandadults[29].
Mostmeningiomashavesimplegenomes,withfewermutations,rearrangementsandcopy-numberalter-ationsthanreportedinotheradulttumors.
However,severalmeningiomasharbormorecomplexpatternsofcopy-numberchangesandrearrangements,includingonetumorwithchromothripsis.
AsubsetofmeningiomaslackingNF2alterationsharboredrecurrentoncogenicmu-tationsinAKT1(p.
Glu17Lys)andSMO(p.
Trp535Leu),andexhibitedimmunohistochemicalevidenceofacti-vationofthesepathways[11].
Inanotherstudy,SMOmutations,whichactivatetheSHHsignalingpathway,wereidentifiedin~5%ofnon-NF2mutantmeningiomas.
Collectively,thesefindingsidentifydistinctmeningiomasubtypes[30].
Moreover,somefindingsprovideara-tionaletoexploretheuseofSMOandBCL2inhibitorsasadjuvanttherapyfortreatmentofDLBCLoftheGCtype[31].
TheexpressionofSMOinNPCisgenerallyhigh,whereasexpressionofPTCH-1isrelativelylow.
DownregulationofPTCH1andupregulationofSMOmaycauseabnormalactivationoftheHHsignalingpathwayinNPC,albeitthatthegenesisanddevelop-mentofNPCmaybeassociatedwithabnormalactiva-tionofHHsignaling[32].
TheHHsignalingpathwayiscontrolledbyPTCH1andSMO,whicharelocatedonthetargetcell'smembrane.
Asthereceptorofsonichedgehog(SHH),thePTCH1gene(51kb)encodesa12-transmembrane-domainprotein(total,1,447aminoacids).
Therearetwohomologousgenesinhumans,PTCH2andPTCH1,whichnegativelyregulateSMO,aG-protein-coupledreceptor,throughSHH.
SMObelongstotheGprotein-coupledreceptorFZ/SMOsuperfamily,containingafieldthatcrossesthecellmembraneseventimes.
Withoutaligand(SHH),PTCHandSMOformaninhibitorycompound,restrainingsignaltransductionthroughouttheentirepathway.
However,whenSHHbindstoPTCH1,SMOescapesfromtheinhibitionbyPTCH1,resultinginenhancedexpressionofdownstreamtargetgenes,includingtheGLIsuperfamily[33].
Asshownbynumerousresearchers,abnormalactivationoftheHHsignalingpathwayiscloselyrelatedtothedevelopmentofavarietyoftumors,andbothPTCH1andSMOplayacriticalroleinthispathway.
TheHHsignalingcascadeishighlyconservedandinvolvedinthedevelopmentofdiseasethroughoutevolution.
Nevertheless,comparedwithotherpathways,ourmechanisticunderstandingofHHsignaltransductionisremarkablyincomplete.
Intheabsenceofligand,theHHreceptorPatched(Ptc),repressesthekeysignaltransducerSmoothened(Smo)throughanunknownmechanism.
HHbindingtoPtcalleviatesthisrepression,causingSmoredistributiontotheplasmamembrane,phosphorylationandsubsequentopeningoftheSmocyto-plasmictailandSmooligomerization.
However,theorderandinterdependenceoftheseeventsarepoorlyunder-stood.
WehavemathematicallymodeledandsimulatedSmoactivationfortwoalternativemodesofactivation,withPtcprimarilyaffectingeitherSmolocalizationorphosphorylation.
Here,weshowthatSmolocalizationtotheplasmamembraneissufficientforphosphorylationofthecytoplasmictailinthepresenceofPtc.
Usingfluor-escencecross-correlationspectroscopy(FCCS),wealsodemonstratedthatinactivationofPtcbyHHinducesSmoclusteringirrespectiveofSmophosphorylation.
Ourob-servationsthereforesupportamodelofHHsignaltrans-ductionwherebysubcellularlocalizationofSmo,andnotphosphorylation,istheprimarytargetofPtcfunction[31].
Severalstudies[34-36]haveconfirmedthatthePTCH1geneisatumorsuppressor;thissuggeststhatitsmutationincreasesthelikelihoodofdevelopingcancer,althoughfewreportsregardingtherelationshipbetweenSMOandKCOThavebeenpublished.
SMOmutationsCodingvariantsofSMOwereidentifiedfollowingouranalysis,andthemutationsarelistedinTable2.
ThefourSMOmutationsdetectedweredistributedonfiveexons(exons2,3,5,6,and10),ofwhichthereweretwosynonymousmutationsandthreemissensemutations.
Thefirstalterationwaslocatedatnucleotideposition776ofexon2,aheterozygousmutationfromTtoA(Figure1A);thisalterationhasnotyetbeenreportedbyotherresearchers.
Wedeterminedthat9of12KCOTpatientshavethisalteration.
Aftercomparingtheproteinaminoacidsequencesofhuman,rice,mouse,andDros-ophila(Figure2),weconcludedthatthisaminoacidisrelativelyconserved.
Meanwhile,followingGenomicsonlinesoftwareSIFT[37](http://sift.
jcvi.
org/)analysis,weconcludedthattheaminoacidchangeresultsinanegativeanddamagingeffectontheprotein(ProveanPreduction:Ruietal.
Head&FaceMedicine2014,10:36Page3of7http://www.
head-face-med.
com/content/10/1/36Deleterious;SiftPreduction:Damaging).
Throughfurtheranalysis,wediscoveredthattheaminoacidislocatedinacysteine-richdomain(CRD)oftheSmoothenedreceptor(Smo)integralmembraneprotein.
TheCRDisoneofthekeyplayersintheHHsignalingpathway,andiscriticalfordevelopment,cellgrowthandmigration,aswellasstemcellmaintenance.
TheCRDofSmoisconservedinverte-bratesandcanalsobeidentifiedininvertebrates.
Thepre-cisefunctionoftheCRDinSmoisunknown.
MutationsintheDrosophilaCRDdisruptSmoactivityinvivo,whiledeletionoftheCRDinmammaliancellsdoesnotappeartoaffectSmooverexpression[38-40].
Therefore,webe-lievethatthisaminoacidmaytremendouslyimpactdevel-opment,cellgrowthandmigration,aswellasstemcellmaintenance.
Itisimportanttonotethatthepeakofthisalterationisdifferentfrommutationsatotherpositions.
Forexample,thepeakofAisrelativelylowerthanT,butnotashighashalfofT.
Accordingtoarecentreport[41],thismaybeduetotheheterogeneityoftumorsinissueswedetected,indicatingthatsomecellshavemutatedwhileothersremainunchanged.
Thesecondalteration,aheterozygousmutationfromAtoG,waslocatedatnucleotideposition862ofexon3(Figure1B)butdoesnotresultinanaminoacidchangeatp.
G194.
Thethirdmutation(c.
G1444C,withG/Calleles)wasidentifiedinseventumors(Figure1C),butdoesnotresultinanaminoacidchangeatp.
G388.
Oneinterestingfactaboutthissynonymousmutationis,withtheexceptionofonesporadicKCOT,allothersamples(includingsporadicandNBCCS-associated)wereiden-tifiedwiththismutation,ofwhich17werehomozygousand13wereheterozygous.
ThefrequencyofGreachedahighof0.
7581,whichisfarhigherthanthestatisticre-portedinthesingle-nucleotidepolymorphism(SNP)databasecuratedbytheNationalCentreforBiotechnol-ogyInformation(NCBI)andthe1000GenomesProject(minorallelefrequency[MAF]=0.
2277).
Thismutationhasbeenreportedinmanyothercelllinesassociatedwithmesothelioma,suchasMSTO-211H,NCI-H28,JU77,LO68,NO36,ONE58,andSTY51[42].
Therefore,itishighlylikelythatthismutationplaysasignificantroleintheoccurrenceoftumors.
However,whetherthismutationisrelatedtoKCOTsshouldbedetermined.
Oneofthemissensemutations(c.
G2049C,withG/Calleles),whichhasbeenreportedpreviously,hasun-knownfunctionalimplications(Figure1D).
Thismuta-tionleadstoanaminoacidchangeatp.
Ser590Thr;itisimportanttonotethatbothValandGlycanbephos-phorylated.
Meanwhile,SIFTsoftwareanalysisrevealedthataninteractivemutationbetweentheseaminoacidsisneutralandtolerated(Table3).
Infact,thisalteredaminoacidislocatedwithintheintracellularloopsofSmoothened,whichmaybecriticallyrelatedtodown-streamproteins,andsubsequentlyimpactthefunctioningofSMO,therebyimpactingactivationoftheHHsignalingpathway[43].
Themissensemutation(c.
T1281G,withT/Calleles)hasnotbeenreportedpreviously(Figure1E).
Thismu-tationleadstoanaminoacidchange(p.
Val334Gly)lo-catedonthefirstextracellularloopofSmoothened,whichiscloselyassociatedwiththefunctionofSMO.
Functionalstudiesontransmembraneregionsarere-strictedbytechnologicalconditions,butaccordingtotheSIFTsoftware(Table3),mutationofthisaminoacidleadstodamagingchangesintheprotein;therefore,webelievethatthismutationstronglyimpactsdiseaseoccurrence.
AdditionalstudiesarerequiredtodeterminewhetherthismutationcausesfunctionalchangesintheSMOgene.
ThemodeofactionbetweentheSMOandPTCHgeneshasyettobeelucidated,butrecentstudieshaveshownthatanymissinggeneinyeastwillimposepressureonthecelltocompensate,therebyleadingtoadd-itionalgeneticmutations[44].
MostSMOgenemutationsTable2SMOmutationsin12sporadicand19NBCCS-associatedKCOTsHistologySampleIDExon/intronNucleotidedefinitionAminoaciddefinitiondbSNPrs#clusterIDMAFClassFunctionS*K1,K2,K4,K5,K6,K7,K8,K10,K11Exon2c.
T776Ap.
Phe166Ile——MissenseUnknownSK7Exon3c.
A862Gp.
=(194)Glurs563342500.
0266SynonymousPolymorphismSK4Exon5c.
T1281Gp.
Val334Gly——MissenseUnknownSK2,K3,K4,K5,K6,K7,K8,K9,K10,K11,K12Exon6c.
G1444Cp.
=(388)Glyrs22286170.
2277SynonymousPolymorphismN*N1,N2,N3,N4,N5,N6,N7,N8,N9,N10,N11,N12,N13,N14,N15,N16,N17,N18,N19,Exon6c.
G1444Cp.
=(388)Glyrs22286170.
2277SynonymousPolymorphismSK1Exon10c.
G2049Cp.
Ser590Thrrs1144068350.
010MissensePolymorphism*S=sporadic,N=NBCCS-associated.
Ruietal.
Head&FaceMedicine2014,10:36Page4of7http://www.
head-face-med.
com/content/10/1/36detectedareseriousalterations,suchasmissenseandsynonymousmutations,whileframeshiftandnonsensemutationshavenotbeendetected.
Therefore,webelievethattheSMOgenemutationmayserveasadrivingforceinpatientswithKCOTs.
TocompensateforthedefectscausedbytheSMOmutation,PTCH1causesthesamesignalingpathwaytomutate,leadingtomuta-tion(s)inthePTCHgene.
Thisconjectureneedsfurtherworktoconfirm.
Inmostcases,peoplebelievethattumorgrowthisdrivenbythe"latest"cancercellsubsetsbecausetheycarrymostcancermutations.
However,manymutationsexistatalowfrequency,suggestingthattumorscontainmanysubclonestherelationshipsamongwhicharestillunclear.
Arecentreportexaminingthehet-erogeneityofbreastcancer[41]showedthatstemcellsinbreastepitheliacoulddifferentiateintoluminalandbasalcells,whichconstitutetheepithelia.
SomealsobelievethatbreastneoplasmsinducedthroughWnt1overexpressionFigure1SMOmissensemutationinsporadicKCOTs.
A.
Thisalternationatnucleotideposition776ofexon2isaheterozygousmutationfromTtoAandcausesthep.
Phe166Ilemutation.
B.
Thisalternationatnucleotideposition862ofexon3isaheterozygousmutationfromAtoG.
However,thisnucleotidechangedoesnotresultinaminoacidchange(p.
=(194)Glu).
Minorallelefrequency(MAF)=0.
0266.
C.
Thisalternationatnucleotideposition1444ofexon6wasidentifiedinseventumors.
Among31patientssequenced,17arehomozygousand13areheterozygousatthissite.
Thisnucleotidechangedoesnotresultinanaminoacidchange(p.
=(388)Gly).
MAF=0.
2277.
D.
Thisalternationatnucleotideposition2049ofexon10isaheterozygousmutationfromGtoCandcausesthep.
Ser590Thrmutation.
E.
Thisalternationatnucleotideposition1281ofexon5isaheterozygousmutationfromTtoCandcausesthep.
Val334Glymutation.
Figure2Conservedaminoacidsinhuman,rice,mouse,andDrosophila.
Aminoacidsequencesbetweenhuman,rice,mouse,andDrosophilaaresimilar(t),indicatingthataminoacidresiduesarerelativelyconservedatthemutationsitedetected(c.
T776A).
Ruietal.
Head&FaceMedicine2014,10:36Page5of7http://www.
head-face-med.
com/content/10/1/36arederivedfromthiscelltype.
Paracrineinteractionbe-tweentwocelltypes,whichisdrivenbysignalingmole-cules,anditsshortdistance,maintainsco-existenceoftwotypesofpedigree.
OnlyluminalcellscanproduceWnt1,whereasbasalcellsrelyonthisproteintoprolifer-ate.
Wnt1inducestheHrasgeneinbasalcellsofbreastneoplasms,whichmaycontainamutationthatdrivescancerprogression,butthismutationhasnotyetbeendetectedincancerouscellsinthelumen.
Thereisco-operationbetweenbasalandluminalcellclones;thiscooperationisnecessaryforWnt1todrivetwotypesofcellsintumors.
Inaddition,KCOTsoccurduringtheearlystagesofdentalepithelialformation,duringwhichdentallaminaanditsresidential,epitheliumandstromainteractwithothersduringgrowth,subsequentlyinducingdifferentiation.
Therefore,wehypothesizethatmutationofsomegenesinthesubcutaneousinterstitialtissuemayimposepressureonepithelialgenestocompensateforthedefect,andthattheSMOmutationmayplayacriticalroleinthisprocess.
However,additionalstudiesareneededforconfirmation.
Asweknow,thehedgehogsignalingpathwayisbeingsuggestedtobeadrugtargetforcancertherapyforitsactivationinhumancancers[45-47].
Therefore,wethinkthetwonewlyidentifiedSMOmutationsdeservetobefurtherinvestigatedfortheirtherapeuticapplicationincancertreatment.
ConclusionsFollowingtheanalysisofSMOgenemutationsandcom-biningourresultswithsimilarstudies,weconcludethatSMOplaysanimportantroleintheHHsignalingpathwayandmayberesponsibleforthedevelopmentofKCOTsandNBCCS.
However,furtherresearchfocusingonthemechanismoftheirinfluenceontheSHHsignalingpathwayisrequired.
CompetinginterestsTheauthorsdeclarethattheyhavenocompetinginterests.
Authors'contributionsFCconceivedanddesignedtheresearch.
JFQandYYHprovidedsamples.
RZandLYPperformedtheexperiments,andcollected,analyzedandinterpretedthedata.
LYPwrotethemanuscript,andeditedtablesandfigures.
RZeditedthemanuscript,tablesandfigurelegends.
FCandTJLreviewedandeditedthemanuscript.
Allauthorsreadandapprovedthefinalmanuscript.
AcknowledgmentsWegratefullyacknowledgethepatientswhoparticipatedinthisstudyandDr.
QianZhangforhertechnicalassistance.
ThisstudywassupportedbygrantsfromtheNationalNaturalScienceFoundationofChina(nos.
81030018,30872900and81200762).
TheEnglishinthisdocumenthasbeencheckedbyatleasttwoprofessionaleditors,bothnativespeakersofEnglish.
Foracertificate,pleasesee:http://www.
textcheck.
com/certificate/GilDRH.
Received:18June2014Accepted:29August2014Published:5September2014References1.
BhargavaD,DeshpandeA,PogrelMA:Keratocysticodontogenictumour(KCOT)–acysttoatumour.
OralMaxillofacSurg2012,16(2):163–170.
2.
NayakMT,SinghA,SinghviA,SharmaR:Odontogenickeratocyst:whatisinthenameJNatScBiolMed2013,4(2):282–285.
3.
LiTJ:Theodontogenickeratocyst:acyst,oracysticneoplasmJDentRes2011,90(2):133–142.
4.
GuXM,ZhaoHS,SunLS,LiTJ:PTCHmutationsinsporadicandgorlin-syndrome-relatedodontogenickeratocysts.
JDentRes2006,85(9):859–863.
5.
GuoYY,ZhangJY,LiXF,LuoHY,ChenF,LiTJ:PTCH1genemutationsinkeratocysticodontogenictumors:astudyof43Chinesepatientsandasystematicreview.
PLoSONE2013,8(10):e77305.
6.
PanS,DongQ,SunLS,LiTJ:MechanismsofinactivationofPTCH1geneinnevoidbasalcellcarcinomasyndrome:modificationofthetwo-hithypothesis.
ClinCancerRes2010,16(2):442–450.
7.
LiTJ,YuanJW,GuXM,SunLS,ZhaoHS:PTCHgermlinemutationsinChinesenevoidbasalcellcarcinomasyndromepatients.
OralDis2008,14(2):174–179.
8.
MusaniV,SabolM,CarD,OzreticP,KalafaticD,MauracI,OreskovicS,LevanatS:PTCH1genepolymorphismsinovariantumors:potentialprotectiveroleofc.
3944Tallele.
Gene2013,517(1):55–59.
9.
HahnH,WickingC,ZaphiropoulousPG,GailaniMR,ShanleyS,ChidambaramA,VorechovskyI,HolmbergE,UndenAB,GilliesS,HahnH,WickingC,ZaphiropoulousPG,GailaniMR,ShanleyS,ChidambaramA,VorechovskyI,HolmbergE,UndenAB,GilliesS,NegusK,SmythI,PressmanC,LeffellDJ,GerrardB,GoldsteinAM,DeanM,ToftgardR,Chenevix-TrenchG,WainwrightB,BaleAE:MutationsofthehumanhomologofDrosophilapatchedinthenevoidbasalcellcarcinomasyndrome.
Cell1996,85(6):841–851.
10.
StoneDM,HynesM,ArmaniniM,SwansonTA,GuQ,JohnsonRL,ScottMP,PennicaD,GoddardA,PhillipsH,NollM,HooperJE,deSauvageF,RosenthalA:Thetumour-suppressorgenepatchedencodesacandidatereceptorforSonichedgehog.
Nature1996,384(6605):129–134.
11.
BrastianosPK,HorowitzPM,SantagataS,JonesRT,McKennaA,GetzG,LigonKL,PalescandoloE,VanHummelenP,DucarMD,RazaA,SunkavalliA,MacconaillLE,Stemmer-RachamimovAO,LouisDN,HahnWC,DunnIF,BeroukhimR:GenomicsequencingofmeningiomasidentifiesoncogenicSMOandAKT1mutations.
NatGenet2013,45(3):285–289.
12.
WangC,WuH,KatritchV,HanGW,HuangXP,LiuW,SiuFY,RothBL,CherezovV,StevensRC:Structureofthehumansmoothenedreceptorboundtoanantitumouragent.
Nature2013,497(7449):338–343.
13.
RuatM,HochL,FaureH,RognanD:Structureofthesmoothenedreceptor.
MedSci2013,29(10):855–860.
14.
JiangJ,HuiCC:Hedgehogsignalingindevelopmentandcancer.
DevCell2008,15(6):801–812.
Table3PROVEANhumanproteinbatchresultVariationProteinsequencechangePROVEANpredictionSiftpredictionRow_No.
Protein_IDPositionResidue_REFResidue_ALTScorePrediction(cutoff=2.
5)ScorePrediction(cutoff=0.
05)1NP_005622.
1166FI5.
385Deleterious0.
002Damaging2NP_005622.
1334VG5.
063Deleterious0.
003Damaging3NP_005622.
1590ST1.
487Neutral0.
219ToleratedChangesataminoacids166and344causeadeleteriousalterationandadamagingresult.
Thechangeataminoacidposition590isrelativelyneutralandtoleratedbytheprotein.
Ruietal.
Head&FaceMedicine2014,10:36Page6of7http://www.
head-face-med.
com/content/10/1/3615.
AmakyeD,JaganiZ,DorschM:UnravelingthetherapeuticpotentialoftheHedgehogpathwayincancer.
NatMed2013,19(11):1410–1422.
16.
YangW,WangJ,MooreDC,LiangH,DoonerM,WuQ,TerekR,ChenQ,EhrlichMG,QuesenberryPJ,NeelBG:Ptpn11deletioninanovelprogenitorcausesmetachondromatosisbyinducinghedgehogsignalling.
Nature2013,499(7459):491–495.
17.
RenY,CowanRG,MigoneFF,QuirkSM:Overactivationofhedgehogsignalingaltersdevelopmentoftheovarianvasculatureinmice.
BiolReprod2012,86(6):174.
18.
JorgensenTJ,RuczinskiI,YaoShugartY,WhelessL,BerthierSchaadY,KessingB,Hoffman-BoltonJ,HelzlsouerKJ,KaoWH,FrancisL,AlaniRM,StricklandPT,SmithMW,AlbergAJ:Apopulation-basedstudyofhedgehogpathwaygenevariantsinrelationtothedualriskofbasalcellcarcinomaplusanothercancer.
CancerEpidemiol2012,36(5):e288–e293.
19.
YangC,ChenW,ChenY,JiangJ:SmoothenedtransducesHedgehogsignalbyformingacomplexwithEvc/Evc2.
CellRes2012,22(11):1593–1604.
20.
ShenF,ChengL,DouglasAE,RioboNA,ManningDR:SmoothenedisafullycompetentactivatoroftheheterotrimericGproteinG(i).
MolPharmacol2013,83(3):691–697.
21.
MyersBR,SeverN,ChongYC,KimJ,BelaniJD,RychnovskyS,BazanJF,BeachyPA:Hedgehogpathwaymodulationbymultiplelipidbindingsitesonthesmoothenedeffectorofsignalresponse.
DevCell2013,26(4):346–357.
22.
MaoJ,FanPH,MaW,ZhangQQ,WangB,FanSJ,LiLH:Down-regulationofSmoothenedgeneexpressioninhibitsproliferationofbreastcancerstemcells.
ZhonghuaBingLiXueZaZhi/ChineseJournalofPathology2013,42(4):262–266.
23.
RuatM,HochL,FaureH,RognanD:TargetingofSmoothenedfortherapeuticgain.
TrendsPharmacolSci2014,35(5):237–246.
24.
ThompsonL:WorldHealthOrganizationclassificationoftumours:pathologyandgeneticsofheadandnecktumours.
EarNoseThroatJ2006,85(2):74.
25.
SunLS,LiXF,LiTJ:PTCH1andSMOgenealterationsinkeratocysticodontogenictumors.
JDentRes2008,87(6):575–579.
26.
ZhangX,HarringtonN,MoraesRC,WuMF,HilsenbeckSG,LewisMT:CyclopamineinhibitionofhumanbreastcancercellgrowthindependentofSmoothened(Smo).
BreastCancerResTreat2009,115(3):505–521.
27.
ZhangL,SunZJ,ChenXM,ChenZ:ImmunohistochemicalexpressionofSHH,PTC,SMOandGLI1inglandularodontogeniccystsanddentigerouscysts.
OralDis2010,16(8):818–822.
28.
SweeneyRT,McClaryAC,MyersBR,BiscochoJ,NeahringL,KweiKA,QuK,GongX,NgT,JonesCD,VarmaS,OdegaardJI,SugiyamaT,KoyotaS,RubinBP,TroxellML,PelhamRJ,ZehnderJL,BeachyPA,PollackJR,WestRB:IdentificationofrecurrentSMOandBRAFmutationsinameloblastomas.
NatGenet2014,46(7):722–5.
29.
:GeneticmutationspredictSMOinhibitorresponseinSHHmedulloblastoma.
Cancdiscov2014,4(5):509.
30.
ClarkVE,Erson-OmayEZ,SerinA,YinJ,CotneyJ,OzdumanK,AvsarT,LiJ,MurrayPB,HenegariuO,YilmazS,GünelJM,Carrión-GrantG,YilmazB,GradyC,TanrikuluB,BakircioluM,KaymakalanH,CaglayanAO,SencarL,CeyhunE,AtikAF,BayriY,BaiH,KolbLE,HebertRM,OmaySB,Mishra-GorurK,ChoiM,OvertonJD,HollandEC,etal:Genomicanalysisofnon-NF2meningiomasrevealsmutationsinTRAF7,KLF4,AKT1,andSMO.
Science2013,339(6123):1077–1080.
31.
KunkallaKLY,QuC,LeventakiV,AgarwalNK,SinghRR,VegaF:FunctionalinhibitionofBCL2isneededtoincreasethesusceptibilitytoapoptosistoSMOinhibitorsindiffuselargeB-celllymphomaofgerminalcentersubtype.
AnnHematol2013,92:777–787.
32.
XiaoBY,DangH,GanJY,CaiQ,ZhangGP,ChangH:ExpressionofPTCH-1andSMOmRNAinnasopharyngealcarcinoma.
XiBaoYuFenZiMianYiXueZaZhi2010,26(10):955–958.
33.
AlcedoJ,AyzenzonM,VonOhlenT,NollM,HooperJE:TheDrosophilasmoothenedgeneencodesaseven-passmembraneprotein,aputativereceptorforthehedgehogsignal.
Cell1996,86(2):221–232.
34.
LevanatS,KapplerR,HemmerleinB,DoringP,MusaniV,KomarA,OreskovicS,PavelicB,HahnH:AnalysisofthePTCH1signalingpathwayinovariandermoids.
IntJMolMed2004,14(5):793–799.
35.
HonmaM,OhishiY,UeharaJ,IbeM,KinouchiM,Ishida-YamamotoA,IizukaH:AnovelPTCH1mutationinapatientofnevoidbasalcellcarcinomasyndrome.
JDermatolSci2008,50(1):73–75.
36.
HimeGR,LadaH,FietzMJ,GilliesS,PassmoreA,WickingC,WainwrightBJ:FunctionalanalysisinDrosophilaindicatesthattheNBCCS/PTCH1mutationG509Vresultsinactivationofsmoothenedthroughadominant-negativemechanism.
DevDyn2004,229(4):780–790.
37.
HuJ,NgPC:SIFTindel:predictionsforthefunctionaleffectsofaminoacidinsertions/deletionsinproteins.
PLoSONE2013,8(10):e77940.
38.
Marchler-BauerA,LuS,AndersonJB,ChitsazF,DerbyshireMK,DeWeese-ScottC,FongJH,GeerLY,GeerRC,GonzalesNR,etal:CDD:aconserveddomaindatabaseforthefunctionalannotationofproteins.
NucleicAcidsRes2011,39(Databaseissue):D225–D229.
39.
Marchler-BauerA,AndersonJB,ChitsazF,DerbyshireMK,DeWeese-ScottC,FongJH,GeerLY,GeerRC,GonzalesNR,GwadzM,HeS,HurwitzDI,JacksonJD,KeZ,LanczyckiCJ,LiebertCA,LiuC,LuF,LuS,MarchlerGH,MullokandovM,SongJS,TasneemA,ThankiN,YamashitaRA,ZhangD,ZhangN,BryantSH:CDD:specificfunctionalannotationwiththeconserveddomaindatabase.
NucleicAcidsRes2009,37(Databaseissue):D205–D210.
40.
Marchler-BauerA,BryantSH:CD-Search:proteindomainannotationsonthefly.
NucleicAcidsRes2004,32(WebServerissue):W327–W331.
41.
ClearyAS,LeonardTL,GestlSA,GuntherEJ:TumourcellheterogeneitymaintainedbycooperatingsubclonesinWnt-drivenmammarycancers.
Nature2014,508(7494):113–117.
42.
LimCB,PreleCM,CheahHM,ChengYY,KlebeS,ReidG,WatkinsDN,BalticS,ThompsonPJ,MutsaersSE:Mutationalanalysisofhedgehogsignalingpathwaygenesinhumanmalignantmesothelioma.
PLoSONE2013,8(6):e66685.
43.
CandaceE,CarrollSM,StewartDP,XiaoxiOuyangJ,OgdenSK:TheextracellularloopsofSmoothenedplayaregulatoryroleincontrolofHedgehogpathwayactivation.
Development2012,139(3):612–621.
44.
TengX,Dayhoff-BranniganM,ChengWC,GilbertCE,SingCN,DinyNL,WheelanSJ,DunhamMJ,BoekeJD,PinedaFJ,HardwickJM:Genome-wideconsequencesofdeletinganysinglegene.
MolCell2013,52(4):485–494.
45.
OnishiH,KatanoM:Hedgehogsignalingpathwayasatherapeutictargetinvarioustypesofcancer.
CancerSci2011,102(10):1756–1760.
46.
AbidiA:Hedgehogsignalingpathway:anoveltargetforcancertherapy:vismodegib,apromisingtherapeuticoptionintreatmentofbasalcellcarcinomas.
IndianJPharmacol2014,46(1):3–12.
47.
MatsushitaS,OnishiH,NakanoK,NagamatsuI,ImaizumiA,HattoriM,OdaY,TanakaM,KatanoM:Hedgehogsignalingpathwayisapotentialtherapeutictargetforgallbladdercancer.
CancerSci2014,105(3):272–280.
doi:10.
1186/1746-160X-10-36Citethisarticleas:Ruietal.
:Smoothenedgenealterationsinkeratocysticodontogenictumors.
Head&FaceMedicine201410:36.
SubmityournextmanuscripttoBioMedCentralandtakefulladvantageof:ConvenientonlinesubmissionThoroughpeerreviewNospaceconstraintsorcolorgurechargesImmediatepublicationonacceptanceInclusioninPubMed,CAS,ScopusandGoogleScholarResearchwhichisfreelyavailableforredistributionSubmityourmanuscriptatwww.
biomedcentral.
com/submitRuietal.
Head&FaceMedicine2014,10:36Page7of7http://www.
head-face-med.
com/content/10/1/36
ToassesspotentialDNAalterationsrelatedtokeratocysticodontogenictumors(KCOTs),wesequencedsmoothened(SMO)genesin12sporadicKCOTs.
Methods:Polymerasechainreaction(PCR),capillaryelectrophoresisanddideoxychain-terminationsequencingwereusedtoexaminepotentialDNAalterationsinsporadicKCOTs.
Results:FivealterationsinSMOgenesweredetected.
Fourofthesemutationsconsistedoftwosynonymousandthreemissensemutations;twoofwhichhavenotbeenreportedtodate(c.
T776A,c.
T1281G).
Conclusions:SMOgenesmayplayanimportantroleinthesonichedgehog(SHH)pathwayandcouldalsoberesponsibleforgeneratingKCOTsandNBCCS.
However,theirinfluenceonSHHsignalingremainstobeelucidated.
Keywords:Keratocysticodontogenictumor(KCOT),Hedgehog(HH)signalingpathway,Genemutation,SMOgeneBackgroundOdontogenickeratocyst(OKC)isanaggressive,cysticjawlesionwithstronggrowthpotentialandahighrecurrencerate.
Inrecentyears,theWorldHealthOrganization(WHO)reviseditsnametokeratocysticodontogenictumor(KCOT).
Thisreclassificationisbasedonitsaggressivebehaviorandhighrecurrencerate,emphasizingthatKCOTisabenigntumorratherthanacyst[1-3].
Althoughthegreatmajorityofkerato-cystsoccurinisolationassingle,non-syndromiccysts,theymayalsopresentasmultiplecystsasafeatureofthenevoidbasalcellcarcinomasyndrome(Gorlinsyndrome,OMIM#109400)[4].
Atpresent,therearemanymanuscriptsthatfocusontherelationshipbetweenKCOTandPTCH1(patched)genemutations,demonstratingthatPTCH1,thegeneresponsibleforNBCCS,mayalsoplayanimportantroleinsporadicKCOTs[5-8].
ThePTCH1geneisatumorsuppressorgenelocatedat9q22.
32[9].
Astudyof14patientswithNBCCS-associatedKCOTsand29patientswithsporadicKCOTsindicatedthatmutationsintrans-membrane2(TM2)arecloselyrelatedtothedevelopmentofsporadicKCOTs[5].
Thehedgehog(HH)signalingpathwayisakeyregulatorofembryonicdevelopment,controllingbothcellularpro-liferationandcellfate.
Bindingofsonichedgehog(SHH)toitsreceptor,patched(PTCH1),isbelievedtorelievenormalinhibitionbyPTCH1ofsmoothened(SMO),aseven-spantransmembraneproteinwithhomologytoaG-protein-coupledreceptor[10].
SMOisatumor-relatedgenelocatedat7q32.
3,con-tains12exonsspanningapproximately24kb,anden-codesa787-amino-acidtransmembraneglycoprotein[11].
ItsreceptorisaGprotein-coupledreceptorthatinteractswithPatched,animportantpartoftheHHsignalingpathwayduringembryogenesisaswellasadult-hood[12,13].
TheHHpathwayhasbeendemonstratedtoplayanimportantroleindifferentdevelopment-relatedcancers[14-18],buttheexactmechanismofactionhasnotyetbeenelucidated.
TheproteingeneratedbySMOisdownstreamofPTCH1;thatis,theexpressionofPTCH1restrainstheactivationofSMO,andtherebyinhibitsactivationoftheHHpathway[19-22].
RecentstudieshavehighlightedthetherapeuticvalueofSMO*Correspondence:moleculecf@gmail.
com;litiejun22@vip.
sina.
comEqualcontributors2CentralLaboratory,PekingUniversitySchoolandHospitalofStomatology,22ZhongguancunAvenueSouth,HaidianDistrict,Beijing100081,China1DepartmentofOralPathology,PekingUniversitySchoolandHospitalofStomatology,22ZhongguancunAvenueSouth,HaidianDistrict,Beijing100081,ChinaHEAD&FACEMEDICINE2014Ruietal.
;licenseeBioMedCentralLtd.
ThisisanOpenAccessarticledistributedunderthetermsoftheCreativeCommonsAttributionLicense(http://creativecommons.
org/licenses/by/4.
0),whichpermitsunrestricteduse,distribution,andreproductioninanymedium,providedtheoriginalworkisproperlycredited.
TheCreativeCommonsPublicDomainDedicationwaiver(http://creativecommons.
org/publicdomain/zero/1.
0/)appliestothedatamadeavailableinthisarticle,unlessotherwisestated.
Ruietal.
Head&FaceMedicine2014,10:36http://www.
head-face-med.
com/content/10/1/36antagonistsforthetreatmentofHH-linkedcancers[22,23].
SMO,themainactivatoroftheHHpathwaymayserveasacatalystduringthegenerationofcysts,andtherefore,geneticmutationsofSMOareofgreatimportance.
MethodsTumorsamplesandclinicalbackgroundFourteenKCOTsampleswithadefinitediagnosiswereacquiredfromclinicalsourcesatPekingUniversitySchoolofStomatology,OralandMaxillofacialSurgeryDepartment.
DiagnoseswerebasedonWHOclassifica-tionoftumors:pathologyandgeneticsoftumorsoftheheadandneck[24].
AllsampleswerefromChinesepa-tients(eightmalesandsixfemales).
Agesvariedfrom10to58years,withanaverageof29.
2years.
ExperimentalprotocolsusedinthisstudywerereviewedandapprovedbytheEthicsCommitteeofthePekingUniversityHealthScienceCenter(Peking,China).
Informedconsentwasobtainedfromallsubjects.
DNAisolationandmutationanalysisGenomicDNAsfromepithelialcellsandinterstitialcellsoftumorswereisolatedusingaQIAampDNAMinikit(Qiagen,Hilden,American)accordingtothemanu-facturer'sinstructions.
DNAwasquantifiedusingaNanodrop(ThermoFischer,Wilmington,DE).
SMO(NM_005631.
4)codingsequencesweredeterminedusingpolymerasechainreaction(PCR)and1%agarosegelelectrophoresis.
ThirteenpairsofprimerscoveringtheentirecodingsequencesandafewnucleotidesintotheintronsequencesonbothendsofSMOarede-scribedinTable1.
PCRswereperformedonaPTC-100(MJResearch,Watertown,MA,USA)inafinalvolumeof30μLcontainingapproximately100-ngtemplateDNA,200μmol/LdNTPs,0.
2–0.
4μmol/Leachprimer,1.
25UExTaqpolymerase(TaKaRa,Dalian,China),and10XPCRbuffer(50mmol/LKCl,10mmol/LTris–HCl,pH9.
0,and1.
5mmol/LMgCl2).
Exon1,exon12aandexon12boftheSMOgenewereamplifiedbyatouch-down(TD)PCRtechnique(Table1).
Forexample,TD65-56°Cwasperformedasfollows:aninitialdenaturationstepat95°Cfor5min,followedby40cyclesat94°Cfor30s,30satanannealingtemperaturedecreasingfrom65°Cto56°C(decreaseof1°Ceach2cycles,20cyclesat55°C),30sat72°C,andafinalextensionstepat72°Cfor10min.
Thespecificityofamplifiedproductswasconfirmedby2%agarosegelelectrophoresis.
PCRproductswerepurifiedbyLifeTechnologies(AppliedBiosystems,USA)andsequencedbyanABI3730xlDNAsequencer(AppliedBiosystems,USA).
Bothforwardandreversestrandsweresequenced.
Anymutationde-tectedwasconfirmedusingbothforwardandreverseprimers.
StatisticalanalysisExperimentaldatawereanalyzedusingtheSPSSver.
10.
0softwareandarepresentedasmeans±standarddeviation(SD)usingstatisticalmethodssuchasat-test,analysisofbivariatecorrelation,etc.
APvalue<0.
05wasconsideredtoindicatestatisticalsignificance.
Resultsarerepresenta-tiveoftwoindependentexperiments.
ResultsanddiscussionHedgehog(HH)signalingpathwayHedgehog(HH)isasignaltransductionpathwaycloselyrelatedtocellgrowthanddifferentiation,andplaysavitalroleinembryonicdevelopment.
MutationorabnormalTable1PCRconditionsofSMOexonsExonSize(bp)Forwardprimersequence(5′to3′)Reverseprimersequence(5′to3′)ReferenceAnnealingtemperature(°C)1391GGGCTGCTGCTGCTGCTGGTCCCCGCCCTCTCCAAAACTSunetal.
,2008[25]TD60~502283AAGAACTGTCCTGCCCAGATGCCACTGGACCCTGCCCTATACWangetal.
,2013[12]633305AATAATTTGCCAAGCCAGCCCTTCTGATCATGACCCTTCCCWangetal.
,2013[12]634256AGGGTCATGATCAGAAGGGTCAGTATGCAGTAGGGCAGAGCCWangetal.
,2013[12]635302CTGACTTCTGGGAACCTCCAGGACAGAAGGTGGGTTACTGGCWangetal.
,2013[12]636206GTGGCGCAGGTATAGTGACTGGCCCTATAGGAGCTAGCTGGGWangetal.
,2013[12]637175GACTCCAGAGCCTTAGGACCCTCCCTATGGCTAACTTGTCCCWangetal.
,2013[12]638196AAGCAGTTCTTGGACTGAGCCCCATCCATTGAATCTGCTGTCWangetal.
,2013[12]559379AGTTGGAAGCTGCAGTGGGCAAGGCTGTGCTAGAGGCAGWangetal.
,2013[12]6310228CTCTGGAAAGAATGGCATCGTTCCAAATAATCTGTGTGCCCWangetal.
,2013[12]5511215AATGGCACTGACTATGGGAGGCCACTCTTCAGATCCTCTGGGWangetal.
,2013[12]6312a292GAGCCAGGGCCCCAGGCTCGTACGCTCCCTGTCGGCAAGAGTSunetal.
,2008[25]TD65-5612b315AGTACCATTCCTCGACTGCCTGGTATTGGTTCCTCTCTTTCCSunetal.
,2008[25]TD65-56Ruietal.
Head&FaceMedicine2014,10:36Page2of7http://www.
head-face-med.
com/content/10/1/36expressionofcomponentsofthispathwaywillleadtovariousdevelopmentaldefectsand/ortumors.
AlteredHHsignalingisimplicatedinthedevelopmentofapproxi-mately20-25%ofallcancers,especiallythoseofsofttissues[26].
ThesefindingsalsosuggestthatproteinsoftheHHsignalingpathwayarepredominantlylocatedwithintheepithelialcomponentsofglandularodontogeniccysts(GOCs)anddentigerouscysts(DCs).
Therefore,theHHsignalingpathwaymayplayanimportantroleintheformationofepitheliallining[27].
ThediscoveryofoncogenicmutationsintheHHandmitogen-activatedproteinkinase(MAPK)pathwaysinover80%ofame-loblastomas,locallydestructiveodontogenictumorsofthejaw,wasreportedin2014bygenomicanalysisofarchivalmaterial.
MutationsinSMOarecommoninameloblastomasofthemaxilla,whereasBRAFmuta-tionsarepredominantintumorsofthemandible[28].
Inaddition,SHHsubgroupmedulloblastomasaregen-eticallydistinctininfants,childrenandadults[29].
Mostmeningiomashavesimplegenomes,withfewermutations,rearrangementsandcopy-numberalter-ationsthanreportedinotheradulttumors.
However,severalmeningiomasharbormorecomplexpatternsofcopy-numberchangesandrearrangements,includingonetumorwithchromothripsis.
AsubsetofmeningiomaslackingNF2alterationsharboredrecurrentoncogenicmu-tationsinAKT1(p.
Glu17Lys)andSMO(p.
Trp535Leu),andexhibitedimmunohistochemicalevidenceofacti-vationofthesepathways[11].
Inanotherstudy,SMOmutations,whichactivatetheSHHsignalingpathway,wereidentifiedin~5%ofnon-NF2mutantmeningiomas.
Collectively,thesefindingsidentifydistinctmeningiomasubtypes[30].
Moreover,somefindingsprovideara-tionaletoexploretheuseofSMOandBCL2inhibitorsasadjuvanttherapyfortreatmentofDLBCLoftheGCtype[31].
TheexpressionofSMOinNPCisgenerallyhigh,whereasexpressionofPTCH-1isrelativelylow.
DownregulationofPTCH1andupregulationofSMOmaycauseabnormalactivationoftheHHsignalingpathwayinNPC,albeitthatthegenesisanddevelop-mentofNPCmaybeassociatedwithabnormalactiva-tionofHHsignaling[32].
TheHHsignalingpathwayiscontrolledbyPTCH1andSMO,whicharelocatedonthetargetcell'smembrane.
Asthereceptorofsonichedgehog(SHH),thePTCH1gene(51kb)encodesa12-transmembrane-domainprotein(total,1,447aminoacids).
Therearetwohomologousgenesinhumans,PTCH2andPTCH1,whichnegativelyregulateSMO,aG-protein-coupledreceptor,throughSHH.
SMObelongstotheGprotein-coupledreceptorFZ/SMOsuperfamily,containingafieldthatcrossesthecellmembraneseventimes.
Withoutaligand(SHH),PTCHandSMOformaninhibitorycompound,restrainingsignaltransductionthroughouttheentirepathway.
However,whenSHHbindstoPTCH1,SMOescapesfromtheinhibitionbyPTCH1,resultinginenhancedexpressionofdownstreamtargetgenes,includingtheGLIsuperfamily[33].
Asshownbynumerousresearchers,abnormalactivationoftheHHsignalingpathwayiscloselyrelatedtothedevelopmentofavarietyoftumors,andbothPTCH1andSMOplayacriticalroleinthispathway.
TheHHsignalingcascadeishighlyconservedandinvolvedinthedevelopmentofdiseasethroughoutevolution.
Nevertheless,comparedwithotherpathways,ourmechanisticunderstandingofHHsignaltransductionisremarkablyincomplete.
Intheabsenceofligand,theHHreceptorPatched(Ptc),repressesthekeysignaltransducerSmoothened(Smo)throughanunknownmechanism.
HHbindingtoPtcalleviatesthisrepression,causingSmoredistributiontotheplasmamembrane,phosphorylationandsubsequentopeningoftheSmocyto-plasmictailandSmooligomerization.
However,theorderandinterdependenceoftheseeventsarepoorlyunder-stood.
WehavemathematicallymodeledandsimulatedSmoactivationfortwoalternativemodesofactivation,withPtcprimarilyaffectingeitherSmolocalizationorphosphorylation.
Here,weshowthatSmolocalizationtotheplasmamembraneissufficientforphosphorylationofthecytoplasmictailinthepresenceofPtc.
Usingfluor-escencecross-correlationspectroscopy(FCCS),wealsodemonstratedthatinactivationofPtcbyHHinducesSmoclusteringirrespectiveofSmophosphorylation.
Ourob-servationsthereforesupportamodelofHHsignaltrans-ductionwherebysubcellularlocalizationofSmo,andnotphosphorylation,istheprimarytargetofPtcfunction[31].
Severalstudies[34-36]haveconfirmedthatthePTCH1geneisatumorsuppressor;thissuggeststhatitsmutationincreasesthelikelihoodofdevelopingcancer,althoughfewreportsregardingtherelationshipbetweenSMOandKCOThavebeenpublished.
SMOmutationsCodingvariantsofSMOwereidentifiedfollowingouranalysis,andthemutationsarelistedinTable2.
ThefourSMOmutationsdetectedweredistributedonfiveexons(exons2,3,5,6,and10),ofwhichthereweretwosynonymousmutationsandthreemissensemutations.
Thefirstalterationwaslocatedatnucleotideposition776ofexon2,aheterozygousmutationfromTtoA(Figure1A);thisalterationhasnotyetbeenreportedbyotherresearchers.
Wedeterminedthat9of12KCOTpatientshavethisalteration.
Aftercomparingtheproteinaminoacidsequencesofhuman,rice,mouse,andDros-ophila(Figure2),weconcludedthatthisaminoacidisrelativelyconserved.
Meanwhile,followingGenomicsonlinesoftwareSIFT[37](http://sift.
jcvi.
org/)analysis,weconcludedthattheaminoacidchangeresultsinanegativeanddamagingeffectontheprotein(ProveanPreduction:Ruietal.
Head&FaceMedicine2014,10:36Page3of7http://www.
head-face-med.
com/content/10/1/36Deleterious;SiftPreduction:Damaging).
Throughfurtheranalysis,wediscoveredthattheaminoacidislocatedinacysteine-richdomain(CRD)oftheSmoothenedreceptor(Smo)integralmembraneprotein.
TheCRDisoneofthekeyplayersintheHHsignalingpathway,andiscriticalfordevelopment,cellgrowthandmigration,aswellasstemcellmaintenance.
TheCRDofSmoisconservedinverte-bratesandcanalsobeidentifiedininvertebrates.
Thepre-cisefunctionoftheCRDinSmoisunknown.
MutationsintheDrosophilaCRDdisruptSmoactivityinvivo,whiledeletionoftheCRDinmammaliancellsdoesnotappeartoaffectSmooverexpression[38-40].
Therefore,webe-lievethatthisaminoacidmaytremendouslyimpactdevel-opment,cellgrowthandmigration,aswellasstemcellmaintenance.
Itisimportanttonotethatthepeakofthisalterationisdifferentfrommutationsatotherpositions.
Forexample,thepeakofAisrelativelylowerthanT,butnotashighashalfofT.
Accordingtoarecentreport[41],thismaybeduetotheheterogeneityoftumorsinissueswedetected,indicatingthatsomecellshavemutatedwhileothersremainunchanged.
Thesecondalteration,aheterozygousmutationfromAtoG,waslocatedatnucleotideposition862ofexon3(Figure1B)butdoesnotresultinanaminoacidchangeatp.
G194.
Thethirdmutation(c.
G1444C,withG/Calleles)wasidentifiedinseventumors(Figure1C),butdoesnotresultinanaminoacidchangeatp.
G388.
Oneinterestingfactaboutthissynonymousmutationis,withtheexceptionofonesporadicKCOT,allothersamples(includingsporadicandNBCCS-associated)wereiden-tifiedwiththismutation,ofwhich17werehomozygousand13wereheterozygous.
ThefrequencyofGreachedahighof0.
7581,whichisfarhigherthanthestatisticre-portedinthesingle-nucleotidepolymorphism(SNP)databasecuratedbytheNationalCentreforBiotechnol-ogyInformation(NCBI)andthe1000GenomesProject(minorallelefrequency[MAF]=0.
2277).
Thismutationhasbeenreportedinmanyothercelllinesassociatedwithmesothelioma,suchasMSTO-211H,NCI-H28,JU77,LO68,NO36,ONE58,andSTY51[42].
Therefore,itishighlylikelythatthismutationplaysasignificantroleintheoccurrenceoftumors.
However,whetherthismutationisrelatedtoKCOTsshouldbedetermined.
Oneofthemissensemutations(c.
G2049C,withG/Calleles),whichhasbeenreportedpreviously,hasun-knownfunctionalimplications(Figure1D).
Thismuta-tionleadstoanaminoacidchangeatp.
Ser590Thr;itisimportanttonotethatbothValandGlycanbephos-phorylated.
Meanwhile,SIFTsoftwareanalysisrevealedthataninteractivemutationbetweentheseaminoacidsisneutralandtolerated(Table3).
Infact,thisalteredaminoacidislocatedwithintheintracellularloopsofSmoothened,whichmaybecriticallyrelatedtodown-streamproteins,andsubsequentlyimpactthefunctioningofSMO,therebyimpactingactivationoftheHHsignalingpathway[43].
Themissensemutation(c.
T1281G,withT/Calleles)hasnotbeenreportedpreviously(Figure1E).
Thismu-tationleadstoanaminoacidchange(p.
Val334Gly)lo-catedonthefirstextracellularloopofSmoothened,whichiscloselyassociatedwiththefunctionofSMO.
Functionalstudiesontransmembraneregionsarere-strictedbytechnologicalconditions,butaccordingtotheSIFTsoftware(Table3),mutationofthisaminoacidleadstodamagingchangesintheprotein;therefore,webelievethatthismutationstronglyimpactsdiseaseoccurrence.
AdditionalstudiesarerequiredtodeterminewhetherthismutationcausesfunctionalchangesintheSMOgene.
ThemodeofactionbetweentheSMOandPTCHgeneshasyettobeelucidated,butrecentstudieshaveshownthatanymissinggeneinyeastwillimposepressureonthecelltocompensate,therebyleadingtoadd-itionalgeneticmutations[44].
MostSMOgenemutationsTable2SMOmutationsin12sporadicand19NBCCS-associatedKCOTsHistologySampleIDExon/intronNucleotidedefinitionAminoaciddefinitiondbSNPrs#clusterIDMAFClassFunctionS*K1,K2,K4,K5,K6,K7,K8,K10,K11Exon2c.
T776Ap.
Phe166Ile——MissenseUnknownSK7Exon3c.
A862Gp.
=(194)Glurs563342500.
0266SynonymousPolymorphismSK4Exon5c.
T1281Gp.
Val334Gly——MissenseUnknownSK2,K3,K4,K5,K6,K7,K8,K9,K10,K11,K12Exon6c.
G1444Cp.
=(388)Glyrs22286170.
2277SynonymousPolymorphismN*N1,N2,N3,N4,N5,N6,N7,N8,N9,N10,N11,N12,N13,N14,N15,N16,N17,N18,N19,Exon6c.
G1444Cp.
=(388)Glyrs22286170.
2277SynonymousPolymorphismSK1Exon10c.
G2049Cp.
Ser590Thrrs1144068350.
010MissensePolymorphism*S=sporadic,N=NBCCS-associated.
Ruietal.
Head&FaceMedicine2014,10:36Page4of7http://www.
head-face-med.
com/content/10/1/36detectedareseriousalterations,suchasmissenseandsynonymousmutations,whileframeshiftandnonsensemutationshavenotbeendetected.
Therefore,webelievethattheSMOgenemutationmayserveasadrivingforceinpatientswithKCOTs.
TocompensateforthedefectscausedbytheSMOmutation,PTCH1causesthesamesignalingpathwaytomutate,leadingtomuta-tion(s)inthePTCHgene.
Thisconjectureneedsfurtherworktoconfirm.
Inmostcases,peoplebelievethattumorgrowthisdrivenbythe"latest"cancercellsubsetsbecausetheycarrymostcancermutations.
However,manymutationsexistatalowfrequency,suggestingthattumorscontainmanysubclonestherelationshipsamongwhicharestillunclear.
Arecentreportexaminingthehet-erogeneityofbreastcancer[41]showedthatstemcellsinbreastepitheliacoulddifferentiateintoluminalandbasalcells,whichconstitutetheepithelia.
SomealsobelievethatbreastneoplasmsinducedthroughWnt1overexpressionFigure1SMOmissensemutationinsporadicKCOTs.
A.
Thisalternationatnucleotideposition776ofexon2isaheterozygousmutationfromTtoAandcausesthep.
Phe166Ilemutation.
B.
Thisalternationatnucleotideposition862ofexon3isaheterozygousmutationfromAtoG.
However,thisnucleotidechangedoesnotresultinaminoacidchange(p.
=(194)Glu).
Minorallelefrequency(MAF)=0.
0266.
C.
Thisalternationatnucleotideposition1444ofexon6wasidentifiedinseventumors.
Among31patientssequenced,17arehomozygousand13areheterozygousatthissite.
Thisnucleotidechangedoesnotresultinanaminoacidchange(p.
=(388)Gly).
MAF=0.
2277.
D.
Thisalternationatnucleotideposition2049ofexon10isaheterozygousmutationfromGtoCandcausesthep.
Ser590Thrmutation.
E.
Thisalternationatnucleotideposition1281ofexon5isaheterozygousmutationfromTtoCandcausesthep.
Val334Glymutation.
Figure2Conservedaminoacidsinhuman,rice,mouse,andDrosophila.
Aminoacidsequencesbetweenhuman,rice,mouse,andDrosophilaaresimilar(t),indicatingthataminoacidresiduesarerelativelyconservedatthemutationsitedetected(c.
T776A).
Ruietal.
Head&FaceMedicine2014,10:36Page5of7http://www.
head-face-med.
com/content/10/1/36arederivedfromthiscelltype.
Paracrineinteractionbe-tweentwocelltypes,whichisdrivenbysignalingmole-cules,anditsshortdistance,maintainsco-existenceoftwotypesofpedigree.
OnlyluminalcellscanproduceWnt1,whereasbasalcellsrelyonthisproteintoprolifer-ate.
Wnt1inducestheHrasgeneinbasalcellsofbreastneoplasms,whichmaycontainamutationthatdrivescancerprogression,butthismutationhasnotyetbeendetectedincancerouscellsinthelumen.
Thereisco-operationbetweenbasalandluminalcellclones;thiscooperationisnecessaryforWnt1todrivetwotypesofcellsintumors.
Inaddition,KCOTsoccurduringtheearlystagesofdentalepithelialformation,duringwhichdentallaminaanditsresidential,epitheliumandstromainteractwithothersduringgrowth,subsequentlyinducingdifferentiation.
Therefore,wehypothesizethatmutationofsomegenesinthesubcutaneousinterstitialtissuemayimposepressureonepithelialgenestocompensateforthedefect,andthattheSMOmutationmayplayacriticalroleinthisprocess.
However,additionalstudiesareneededforconfirmation.
Asweknow,thehedgehogsignalingpathwayisbeingsuggestedtobeadrugtargetforcancertherapyforitsactivationinhumancancers[45-47].
Therefore,wethinkthetwonewlyidentifiedSMOmutationsdeservetobefurtherinvestigatedfortheirtherapeuticapplicationincancertreatment.
ConclusionsFollowingtheanalysisofSMOgenemutationsandcom-biningourresultswithsimilarstudies,weconcludethatSMOplaysanimportantroleintheHHsignalingpathwayandmayberesponsibleforthedevelopmentofKCOTsandNBCCS.
However,furtherresearchfocusingonthemechanismoftheirinfluenceontheSHHsignalingpathwayisrequired.
CompetinginterestsTheauthorsdeclarethattheyhavenocompetinginterests.
Authors'contributionsFCconceivedanddesignedtheresearch.
JFQandYYHprovidedsamples.
RZandLYPperformedtheexperiments,andcollected,analyzedandinterpretedthedata.
LYPwrotethemanuscript,andeditedtablesandfigures.
RZeditedthemanuscript,tablesandfigurelegends.
FCandTJLreviewedandeditedthemanuscript.
Allauthorsreadandapprovedthefinalmanuscript.
AcknowledgmentsWegratefullyacknowledgethepatientswhoparticipatedinthisstudyandDr.
QianZhangforhertechnicalassistance.
ThisstudywassupportedbygrantsfromtheNationalNaturalScienceFoundationofChina(nos.
81030018,30872900and81200762).
TheEnglishinthisdocumenthasbeencheckedbyatleasttwoprofessionaleditors,bothnativespeakersofEnglish.
Foracertificate,pleasesee:http://www.
textcheck.
com/certificate/GilDRH.
Received:18June2014Accepted:29August2014Published:5September2014References1.
BhargavaD,DeshpandeA,PogrelMA:Keratocysticodontogenictumour(KCOT)–acysttoatumour.
OralMaxillofacSurg2012,16(2):163–170.
2.
NayakMT,SinghA,SinghviA,SharmaR:Odontogenickeratocyst:whatisinthenameJNatScBiolMed2013,4(2):282–285.
3.
LiTJ:Theodontogenickeratocyst:acyst,oracysticneoplasmJDentRes2011,90(2):133–142.
4.
GuXM,ZhaoHS,SunLS,LiTJ:PTCHmutationsinsporadicandgorlin-syndrome-relatedodontogenickeratocysts.
JDentRes2006,85(9):859–863.
5.
GuoYY,ZhangJY,LiXF,LuoHY,ChenF,LiTJ:PTCH1genemutationsinkeratocysticodontogenictumors:astudyof43Chinesepatientsandasystematicreview.
PLoSONE2013,8(10):e77305.
6.
PanS,DongQ,SunLS,LiTJ:MechanismsofinactivationofPTCH1geneinnevoidbasalcellcarcinomasyndrome:modificationofthetwo-hithypothesis.
ClinCancerRes2010,16(2):442–450.
7.
LiTJ,YuanJW,GuXM,SunLS,ZhaoHS:PTCHgermlinemutationsinChinesenevoidbasalcellcarcinomasyndromepatients.
OralDis2008,14(2):174–179.
8.
MusaniV,SabolM,CarD,OzreticP,KalafaticD,MauracI,OreskovicS,LevanatS:PTCH1genepolymorphismsinovariantumors:potentialprotectiveroleofc.
3944Tallele.
Gene2013,517(1):55–59.
9.
HahnH,WickingC,ZaphiropoulousPG,GailaniMR,ShanleyS,ChidambaramA,VorechovskyI,HolmbergE,UndenAB,GilliesS,HahnH,WickingC,ZaphiropoulousPG,GailaniMR,ShanleyS,ChidambaramA,VorechovskyI,HolmbergE,UndenAB,GilliesS,NegusK,SmythI,PressmanC,LeffellDJ,GerrardB,GoldsteinAM,DeanM,ToftgardR,Chenevix-TrenchG,WainwrightB,BaleAE:MutationsofthehumanhomologofDrosophilapatchedinthenevoidbasalcellcarcinomasyndrome.
Cell1996,85(6):841–851.
10.
StoneDM,HynesM,ArmaniniM,SwansonTA,GuQ,JohnsonRL,ScottMP,PennicaD,GoddardA,PhillipsH,NollM,HooperJE,deSauvageF,RosenthalA:Thetumour-suppressorgenepatchedencodesacandidatereceptorforSonichedgehog.
Nature1996,384(6605):129–134.
11.
BrastianosPK,HorowitzPM,SantagataS,JonesRT,McKennaA,GetzG,LigonKL,PalescandoloE,VanHummelenP,DucarMD,RazaA,SunkavalliA,MacconaillLE,Stemmer-RachamimovAO,LouisDN,HahnWC,DunnIF,BeroukhimR:GenomicsequencingofmeningiomasidentifiesoncogenicSMOandAKT1mutations.
NatGenet2013,45(3):285–289.
12.
WangC,WuH,KatritchV,HanGW,HuangXP,LiuW,SiuFY,RothBL,CherezovV,StevensRC:Structureofthehumansmoothenedreceptorboundtoanantitumouragent.
Nature2013,497(7449):338–343.
13.
RuatM,HochL,FaureH,RognanD:Structureofthesmoothenedreceptor.
MedSci2013,29(10):855–860.
14.
JiangJ,HuiCC:Hedgehogsignalingindevelopmentandcancer.
DevCell2008,15(6):801–812.
Table3PROVEANhumanproteinbatchresultVariationProteinsequencechangePROVEANpredictionSiftpredictionRow_No.
Protein_IDPositionResidue_REFResidue_ALTScorePrediction(cutoff=2.
5)ScorePrediction(cutoff=0.
05)1NP_005622.
1166FI5.
385Deleterious0.
002Damaging2NP_005622.
1334VG5.
063Deleterious0.
003Damaging3NP_005622.
1590ST1.
487Neutral0.
219ToleratedChangesataminoacids166and344causeadeleteriousalterationandadamagingresult.
Thechangeataminoacidposition590isrelativelyneutralandtoleratedbytheprotein.
Ruietal.
Head&FaceMedicine2014,10:36Page6of7http://www.
head-face-med.
com/content/10/1/3615.
AmakyeD,JaganiZ,DorschM:UnravelingthetherapeuticpotentialoftheHedgehogpathwayincancer.
NatMed2013,19(11):1410–1422.
16.
YangW,WangJ,MooreDC,LiangH,DoonerM,WuQ,TerekR,ChenQ,EhrlichMG,QuesenberryPJ,NeelBG:Ptpn11deletioninanovelprogenitorcausesmetachondromatosisbyinducinghedgehogsignalling.
Nature2013,499(7459):491–495.
17.
RenY,CowanRG,MigoneFF,QuirkSM:Overactivationofhedgehogsignalingaltersdevelopmentoftheovarianvasculatureinmice.
BiolReprod2012,86(6):174.
18.
JorgensenTJ,RuczinskiI,YaoShugartY,WhelessL,BerthierSchaadY,KessingB,Hoffman-BoltonJ,HelzlsouerKJ,KaoWH,FrancisL,AlaniRM,StricklandPT,SmithMW,AlbergAJ:Apopulation-basedstudyofhedgehogpathwaygenevariantsinrelationtothedualriskofbasalcellcarcinomaplusanothercancer.
CancerEpidemiol2012,36(5):e288–e293.
19.
YangC,ChenW,ChenY,JiangJ:SmoothenedtransducesHedgehogsignalbyformingacomplexwithEvc/Evc2.
CellRes2012,22(11):1593–1604.
20.
ShenF,ChengL,DouglasAE,RioboNA,ManningDR:SmoothenedisafullycompetentactivatoroftheheterotrimericGproteinG(i).
MolPharmacol2013,83(3):691–697.
21.
MyersBR,SeverN,ChongYC,KimJ,BelaniJD,RychnovskyS,BazanJF,BeachyPA:Hedgehogpathwaymodulationbymultiplelipidbindingsitesonthesmoothenedeffectorofsignalresponse.
DevCell2013,26(4):346–357.
22.
MaoJ,FanPH,MaW,ZhangQQ,WangB,FanSJ,LiLH:Down-regulationofSmoothenedgeneexpressioninhibitsproliferationofbreastcancerstemcells.
ZhonghuaBingLiXueZaZhi/ChineseJournalofPathology2013,42(4):262–266.
23.
RuatM,HochL,FaureH,RognanD:TargetingofSmoothenedfortherapeuticgain.
TrendsPharmacolSci2014,35(5):237–246.
24.
ThompsonL:WorldHealthOrganizationclassificationoftumours:pathologyandgeneticsofheadandnecktumours.
EarNoseThroatJ2006,85(2):74.
25.
SunLS,LiXF,LiTJ:PTCH1andSMOgenealterationsinkeratocysticodontogenictumors.
JDentRes2008,87(6):575–579.
26.
ZhangX,HarringtonN,MoraesRC,WuMF,HilsenbeckSG,LewisMT:CyclopamineinhibitionofhumanbreastcancercellgrowthindependentofSmoothened(Smo).
BreastCancerResTreat2009,115(3):505–521.
27.
ZhangL,SunZJ,ChenXM,ChenZ:ImmunohistochemicalexpressionofSHH,PTC,SMOandGLI1inglandularodontogeniccystsanddentigerouscysts.
OralDis2010,16(8):818–822.
28.
SweeneyRT,McClaryAC,MyersBR,BiscochoJ,NeahringL,KweiKA,QuK,GongX,NgT,JonesCD,VarmaS,OdegaardJI,SugiyamaT,KoyotaS,RubinBP,TroxellML,PelhamRJ,ZehnderJL,BeachyPA,PollackJR,WestRB:IdentificationofrecurrentSMOandBRAFmutationsinameloblastomas.
NatGenet2014,46(7):722–5.
29.
:GeneticmutationspredictSMOinhibitorresponseinSHHmedulloblastoma.
Cancdiscov2014,4(5):509.
30.
ClarkVE,Erson-OmayEZ,SerinA,YinJ,CotneyJ,OzdumanK,AvsarT,LiJ,MurrayPB,HenegariuO,YilmazS,GünelJM,Carrión-GrantG,YilmazB,GradyC,TanrikuluB,BakircioluM,KaymakalanH,CaglayanAO,SencarL,CeyhunE,AtikAF,BayriY,BaiH,KolbLE,HebertRM,OmaySB,Mishra-GorurK,ChoiM,OvertonJD,HollandEC,etal:Genomicanalysisofnon-NF2meningiomasrevealsmutationsinTRAF7,KLF4,AKT1,andSMO.
Science2013,339(6123):1077–1080.
31.
KunkallaKLY,QuC,LeventakiV,AgarwalNK,SinghRR,VegaF:FunctionalinhibitionofBCL2isneededtoincreasethesusceptibilitytoapoptosistoSMOinhibitorsindiffuselargeB-celllymphomaofgerminalcentersubtype.
AnnHematol2013,92:777–787.
32.
XiaoBY,DangH,GanJY,CaiQ,ZhangGP,ChangH:ExpressionofPTCH-1andSMOmRNAinnasopharyngealcarcinoma.
XiBaoYuFenZiMianYiXueZaZhi2010,26(10):955–958.
33.
AlcedoJ,AyzenzonM,VonOhlenT,NollM,HooperJE:TheDrosophilasmoothenedgeneencodesaseven-passmembraneprotein,aputativereceptorforthehedgehogsignal.
Cell1996,86(2):221–232.
34.
LevanatS,KapplerR,HemmerleinB,DoringP,MusaniV,KomarA,OreskovicS,PavelicB,HahnH:AnalysisofthePTCH1signalingpathwayinovariandermoids.
IntJMolMed2004,14(5):793–799.
35.
HonmaM,OhishiY,UeharaJ,IbeM,KinouchiM,Ishida-YamamotoA,IizukaH:AnovelPTCH1mutationinapatientofnevoidbasalcellcarcinomasyndrome.
JDermatolSci2008,50(1):73–75.
36.
HimeGR,LadaH,FietzMJ,GilliesS,PassmoreA,WickingC,WainwrightBJ:FunctionalanalysisinDrosophilaindicatesthattheNBCCS/PTCH1mutationG509Vresultsinactivationofsmoothenedthroughadominant-negativemechanism.
DevDyn2004,229(4):780–790.
37.
HuJ,NgPC:SIFTindel:predictionsforthefunctionaleffectsofaminoacidinsertions/deletionsinproteins.
PLoSONE2013,8(10):e77940.
38.
Marchler-BauerA,LuS,AndersonJB,ChitsazF,DerbyshireMK,DeWeese-ScottC,FongJH,GeerLY,GeerRC,GonzalesNR,etal:CDD:aconserveddomaindatabaseforthefunctionalannotationofproteins.
NucleicAcidsRes2011,39(Databaseissue):D225–D229.
39.
Marchler-BauerA,AndersonJB,ChitsazF,DerbyshireMK,DeWeese-ScottC,FongJH,GeerLY,GeerRC,GonzalesNR,GwadzM,HeS,HurwitzDI,JacksonJD,KeZ,LanczyckiCJ,LiebertCA,LiuC,LuF,LuS,MarchlerGH,MullokandovM,SongJS,TasneemA,ThankiN,YamashitaRA,ZhangD,ZhangN,BryantSH:CDD:specificfunctionalannotationwiththeconserveddomaindatabase.
NucleicAcidsRes2009,37(Databaseissue):D205–D210.
40.
Marchler-BauerA,BryantSH:CD-Search:proteindomainannotationsonthefly.
NucleicAcidsRes2004,32(WebServerissue):W327–W331.
41.
ClearyAS,LeonardTL,GestlSA,GuntherEJ:TumourcellheterogeneitymaintainedbycooperatingsubclonesinWnt-drivenmammarycancers.
Nature2014,508(7494):113–117.
42.
LimCB,PreleCM,CheahHM,ChengYY,KlebeS,ReidG,WatkinsDN,BalticS,ThompsonPJ,MutsaersSE:Mutationalanalysisofhedgehogsignalingpathwaygenesinhumanmalignantmesothelioma.
PLoSONE2013,8(6):e66685.
43.
CandaceE,CarrollSM,StewartDP,XiaoxiOuyangJ,OgdenSK:TheextracellularloopsofSmoothenedplayaregulatoryroleincontrolofHedgehogpathwayactivation.
Development2012,139(3):612–621.
44.
TengX,Dayhoff-BranniganM,ChengWC,GilbertCE,SingCN,DinyNL,WheelanSJ,DunhamMJ,BoekeJD,PinedaFJ,HardwickJM:Genome-wideconsequencesofdeletinganysinglegene.
MolCell2013,52(4):485–494.
45.
OnishiH,KatanoM:Hedgehogsignalingpathwayasatherapeutictargetinvarioustypesofcancer.
CancerSci2011,102(10):1756–1760.
46.
AbidiA:Hedgehogsignalingpathway:anoveltargetforcancertherapy:vismodegib,apromisingtherapeuticoptionintreatmentofbasalcellcarcinomas.
IndianJPharmacol2014,46(1):3–12.
47.
MatsushitaS,OnishiH,NakanoK,NagamatsuI,ImaizumiA,HattoriM,OdaY,TanakaM,KatanoM:Hedgehogsignalingpathwayisapotentialtherapeutictargetforgallbladdercancer.
CancerSci2014,105(3):272–280.
doi:10.
1186/1746-160X-10-36Citethisarticleas:Ruietal.
:Smoothenedgenealterationsinkeratocysticodontogenictumors.
Head&FaceMedicine201410:36.
SubmityournextmanuscripttoBioMedCentralandtakefulladvantageof:ConvenientonlinesubmissionThoroughpeerreviewNospaceconstraintsorcolorgurechargesImmediatepublicationonacceptanceInclusioninPubMed,CAS,ScopusandGoogleScholarResearchwhichisfreelyavailableforredistributionSubmityourmanuscriptatwww.
biomedcentral.
com/submitRuietal.
Head&FaceMedicine2014,10:36Page7of7http://www.
head-face-med.
com/content/10/1/36
- genesis777k7.com相关文档
- extracorporeal777k7.com
- 華擎科技股份有限公司九十四報中華民國九十五月刊印政院融監督管委員會指定資訊申報網址:http://mops.tse.com.tw
- arXiv:astro-ph/0107375v1
- 监测777k7.com
- Battery777k7.com
- ij777k7.com
HostYun 新上美国CN2 GIA VPS 月15元
HostYun 商家以前是玩具主机商,这两年好像发展还挺迅速的,有点在要做点事情的味道。在前面也有多次介绍到HostYun商家新增的多款机房方案,价格相对还是比较便宜的。到目前为止,我们可以看到商家提供的VPS主机包括KVM和XEN架构,数据中心可选日本、韩国、香港和美国的多个地区机房,电信双程CN2 GIA线路,香港和日本机房,均为国内直连线路。近期,HostYun上线低价版美国CN2 GIA ...
Megalayer(48元)新增 美国CN2优化线路特价服务器和VPS方案
Megalayer 商家算是新晋的服务商,商家才开始的时候主要是以香港、美国独立服务器。后来有新增菲律宾机房,包括有VPS云服务器、独立服务器、站群服务器等产品。线路上有CN2优化带宽、全向带宽和国际带宽,这里有看到商家的特价方案有增加至9个,之前是四个的。在这篇文章中,我来整理看看。第一、香港服务器系列这里香港服务器会根据带宽的不同区别。我这里将香港机房的都整理到一个系列里。核心内存硬盘IP带宽...

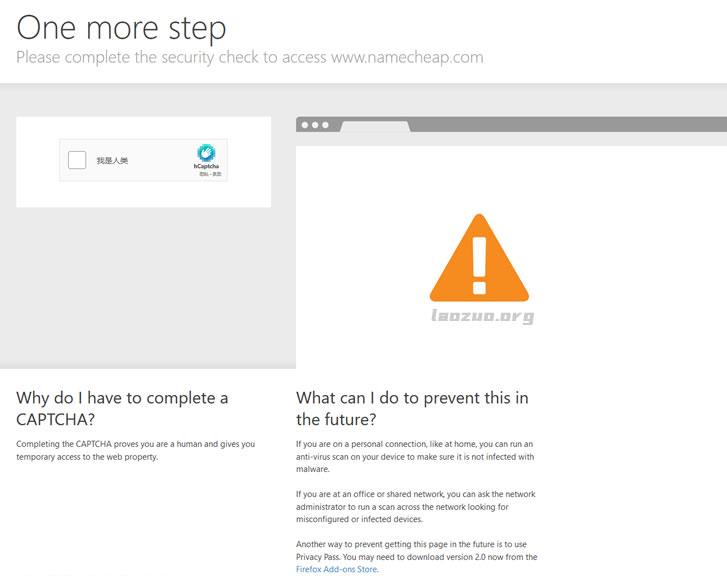
打开海外主机域名商出现"Attention Required"原因和解决
最近发现一个比较怪异的事情,在访问和登录大部分国外主机商和域名商的时候都需要二次验证。常见的就是需要我们勾选判断是不是真人。以及比如在刚才要访问Namecheap检查前几天送给网友域名的账户域名是否转出的,再次登录网站的时候又需要人机验证。这里有看到"Attention Required"的提示。我们只能手工选择按钮,然后根据验证码进行选择合适的标记。这次我要选择的是船的标识,每次需要选择三个,一...
777k7.com为你推荐
-
johncusack谁知道《失控的陪审团》的电影内容是什么?约翰·库萨克在里面演的是什么角色?嘉兴商标注册我想注册个商标怎么注册啊?75ff.com开机出现www.ami.com是什么?怎么解决啊8090lu.com《8090》节目有不有高清的在线观看网站啊?sss17.comwww.com17com.com是什么啊?66smsm.com【回家的欲望(回家的诱惑)大结局】 回家的诱惑全集66 67 68 69 70集QOVD快播观看地址??www.175qq.com这表情是什么?www.1diaocha.com哪个网站做调查问卷可以赚钱 啊铂金血痕手上出现这种血痕是什么情况。有谁知道能告诉下吗? 怎么治疗!铂金血痕求Hp卢修斯,v大,盖特勒重生文,cp不要斯内普和邓不利多,名子和简介就行.最好是晋江的.谢谢.